ZFLNCG03912
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 4.42 |
| SRR891504 | liver | normal | 3.01 |
| SRR516133 | skin | male and 5 month | 2.79 |
| SRR957180 | heart | 7 days after heart tip amputation | 2.75 |
| SRR1188156 | embryo | Control PBS 4 hpi | 2.57 |
| SRR1048059 | pineal gland | light | 2.55 |
| SRR1004788 | larvae | impdh1b morpholino | 2.52 |
| SRR1167756 | embryo | mpeg1 morpholino | 2.36 |
| SRR1188152 | embryo | Insulin 0.5 hpi | 2.28 |
| SRR957181 | heart | normal | 2.18 |
Express in tissues
Correlated coding gene
Download
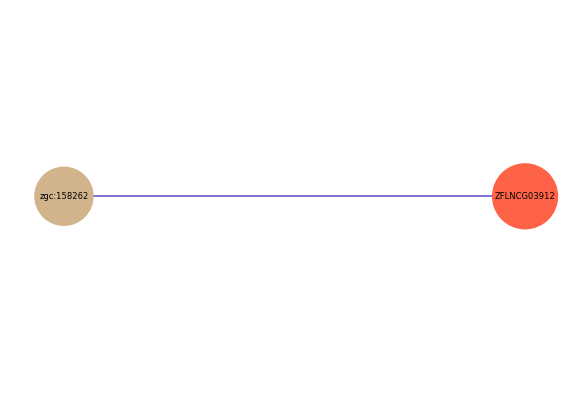
| gene | correlation coefficent |
|---|---|
| zgc:158262 | 0.53 |
Gene Ontology
Gene Ontology of this lncRNA gene hava does not exist!
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03912
TTACTGTACTGGAACCAATGAACTGAAAGGTGTATTTTGTCTGAGACCGCAATGTTTTGAAGTGAGCATT
GACTTCTTTTTTTAACGATTTTAACTTATTTCGCCAGATAGCTATGCATCCTATTCATCTGCTTCGCATT
GTCAGTTCTGTTCGACGCTTATTACAGACTAATAGATGTAGAAGTTTCTCCTCCTGCCAAGTGAGTGCAA
GTTTTTCTGAATTATCAATTTCTCTGACATTCATTCATTTTGTTTTTTCACCATTTTAGCACAACATGGC
ATCTATAAAACACAAAAACGCGAAGAGGATAGAAGGCCTGGACAAGAATGTATGGTAAGAATTGAGAATA
TTTGCTAACACTTTTGTTAATTGTTACACTTAATAATTCAACTTCTACATAAACCATATTTATATTTCAT
GTAAACCATATTTTTGTACATCTCTAAACGTGTATGCAATTTGCATTGTTAACTTTGTTTTCCACTTATT
TAAATGTCCTTCTGTCCTCTTATTCTTATATGTATGCATGCACTCTGAGCTAATGTAAACCCCCAAAAAC
AGCTATATCAGCAAATAAAGCACATTCTAAATATGATAGAATTATGATATGTTTCTTATGTTGTTCAGGG
TGGCTTTCACATCAGTGGCTGCAGATCCTTCTATTGTCAATCTTGGACAGGGTTACCCAGACATTCCTCC
TCCTTCATATGTAAAAGAGGGTCTTGCTCAAGCTGCAATGGTAGACAGGCTGAACCAGTACACACGTGGC
TTTGTATGTTCTGTCTATTCTTCCTAACCATCCTGTAAATAAATACATAAGGCGAACACATAGAAGAACA
ATTTAATCTCTAAACAGCATCACTAGTATTTAATTTCATGCTACTGGTGGCTTAGGTGTAAAATTTAACA
ACACACTCCACAAAACAATATACTATAGCAATGTTGACAGTAATAAGTAAAAGACAATAAATGTTAAAGG
ACATTTAAATATGTTAGGAAAAACACACTTTACACACATGCACAGATAGACAAGTACATAGTCAAAAAAA
GTGTACCCATAGTTTGACTGTCGGTGGAAAATAATTGTAGGCAGTATCGTTTTAAGTAAGGATTTGGTTA
TTGAAAATTGCTAGTGG
TTACTGTACTGGAACCAATGAACTGAAAGGTGTATTTTGTCTGAGACCGCAATGTTTTGAAGTGAGCATT
GACTTCTTTTTTTAACGATTTTAACTTATTTCGCCAGATAGCTATGCATCCTATTCATCTGCTTCGCATT
GTCAGTTCTGTTCGACGCTTATTACAGACTAATAGATGTAGAAGTTTCTCCTCCTGCCAAGTGAGTGCAA
GTTTTTCTGAATTATCAATTTCTCTGACATTCATTCATTTTGTTTTTTCACCATTTTAGCACAACATGGC
ATCTATAAAACACAAAAACGCGAAGAGGATAGAAGGCCTGGACAAGAATGTATGGTAAGAATTGAGAATA
TTTGCTAACACTTTTGTTAATTGTTACACTTAATAATTCAACTTCTACATAAACCATATTTATATTTCAT
GTAAACCATATTTTTGTACATCTCTAAACGTGTATGCAATTTGCATTGTTAACTTTGTTTTCCACTTATT
TAAATGTCCTTCTGTCCTCTTATTCTTATATGTATGCATGCACTCTGAGCTAATGTAAACCCCCAAAAAC
AGCTATATCAGCAAATAAAGCACATTCTAAATATGATAGAATTATGATATGTTTCTTATGTTGTTCAGGG
TGGCTTTCACATCAGTGGCTGCAGATCCTTCTATTGTCAATCTTGGACAGGGTTACCCAGACATTCCTCC
TCCTTCATATGTAAAAGAGGGTCTTGCTCAAGCTGCAATGGTAGACAGGCTGAACCAGTACACACGTGGC
TTTGTATGTTCTGTCTATTCTTCCTAACCATCCTGTAAATAAATACATAAGGCGAACACATAGAAGAACA
ATTTAATCTCTAAACAGCATCACTAGTATTTAATTTCATGCTACTGGTGGCTTAGGTGTAAAATTTAACA
ACACACTCCACAAAACAATATACTATAGCAATGTTGACAGTAATAAGTAAAAGACAATAAATGTTAAAGG
ACATTTAAATATGTTAGGAAAAACACACTTTACACACATGCACAGATAGACAAGTACATAGTCAAAAAAA
GTGTACCCATAGTTTGACTGTCGGTGGAAAATAATTGTAGGCAGTATCGTTTTAAGTAAGGATTTGGTTA
TTGAAAATTGCTAGTGG
