ZFLNCG04043
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 14.22 |
| ERR594417 | retina | normal | 6.00 |
| SRR514028 | retina | control morpholino | 4.72 |
| SRR592699 | pineal gland | normal | 4.14 |
| SRR592701 | pineal gland | normal | 3.77 |
| SRR630464 | 5 dpf | injected epidermidis | 3.75 |
| SRR592703 | pineal gland | normal | 3.66 |
| ERR594416 | retina | transgenic flk1 | 3.59 |
| SRR630463 | 5 dpf | normal | 3.59 |
| SRR1562533 | testis | normal | 3.55 |
Express in tissues
Correlated coding gene
Download
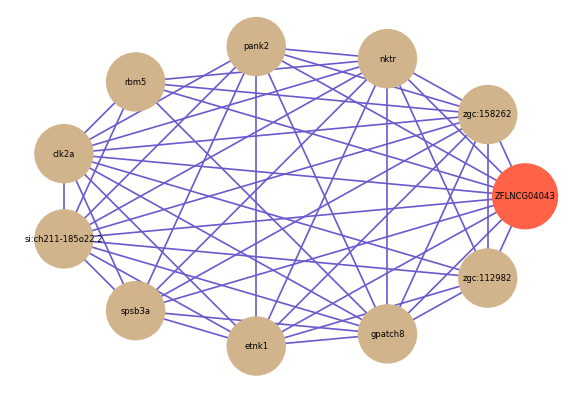
| gene | correlation coefficent |
|---|---|
| zgc:158262 | 0.72 |
| nktr | 0.67 |
| pank2 | 0.63 |
| rbm5 | 0.61 |
| clk2a | 0.61 |
| si:ch211-185o22.2 | 0.60 |
| spsb3a | 0.59 |
| etnk1 | 0.59 |
| gpatch8 | 0.59 |
| zgc:112982 | 0.59 |
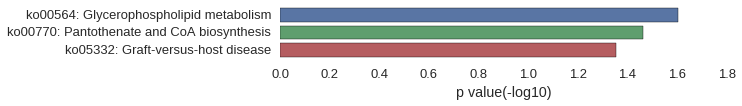
Gene Ontology
Download
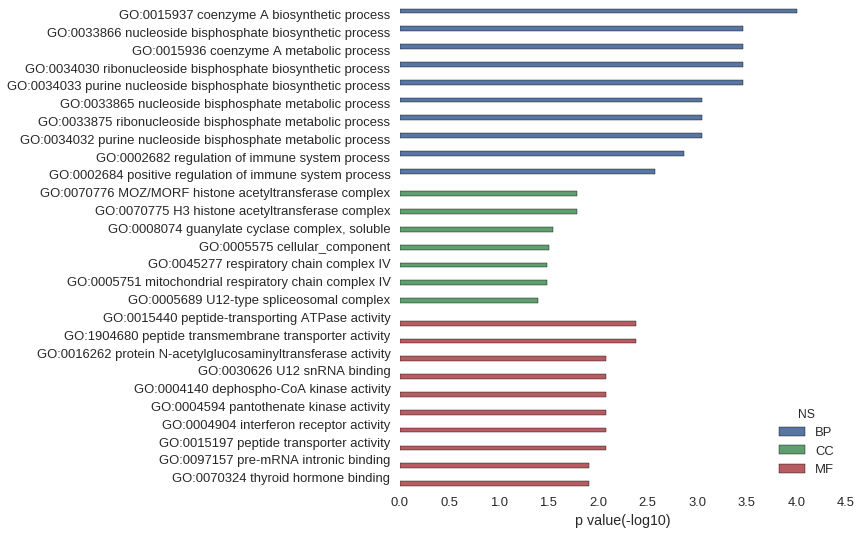
| GO | P value |
|---|---|
| GO:0015937 | 9.96e-05 |
| GO:0033866 | 3.46e-04 |
| GO:0015936 | 3.46e-04 |
| GO:0034030 | 3.46e-04 |
| GO:0034033 | 3.46e-04 |
| GO:0033865 | 8.97e-04 |
| GO:0033875 | 8.97e-04 |
| GO:0034032 | 8.97e-04 |
| GO:0002682 | 1.36e-03 |
| GO:0002684 | 2.64e-03 |
| GO:0070776 | 1.64e-02 |
| GO:0070775 | 1.64e-02 |
| GO:0008074 | 2.85e-02 |
| GO:0005575 | 3.11e-02 |
| GO:0045277 | 3.25e-02 |
| GO:0005751 | 3.25e-02 |
| GO:0005689 | 4.05e-02 |
| GO:0015440 | 4.12e-03 |
| GO:1904680 | 4.12e-03 |
| GO:0016262 | 8.23e-03 |
| GO:0030626 | 8.23e-03 |
| GO:0004140 | 8.23e-03 |
| GO:0004594 | 8.23e-03 |
| GO:0004904 | 8.23e-03 |
| GO:0015197 | 8.23e-03 |
| GO:0097157 | 1.23e-02 |
| GO:0070324 | 1.23e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04043
TAGGATGTGAGCGTAACTCAAAATAAACAAATGCAAAATAAAGATTGAATACAAGTGTATGATGGGAAGT
TTGGATCACTTGGTCTTAATTAAAATCTTACACATTGAGTCCCTGGCATAATTCACTACTGCTGTCCCTG
TGAACTAGACAAACCTACATTGCAGCCATGAATATATAAGAGGCAGCCAAGATTTACTCATTGTTTACCA
GGGTCTCAAGTTAGCTCCACTCATCTCACCTGTCATCTTTTGAGTCCTCAAGTAATTCACTGCCTTTGTC
CTCACGCCACATCTCCATAAATTAAAATGTTACGTGGGCTCTTCCGGTTTAATAATGGAAAATATTAGTA
CATCAGTCTTAAGTCCTCACATTTTCAGTCATCCATTGAGCAATTAATAGCACTATAGGCCAAATATTAA
ACTCGAAGGGTCAATTGTTGGAGTTTTGGTTATTTGCTTTTTGAATTGCTTACTTGGGACCATTGGAGCT
GCCATTTAAGAACTATCTTAGCATTCTACACTGCAAAAAATGC
TAGGATGTGAGCGTAACTCAAAATAAACAAATGCAAAATAAAGATTGAATACAAGTGTATGATGGGAAGT
TTGGATCACTTGGTCTTAATTAAAATCTTACACATTGAGTCCCTGGCATAATTCACTACTGCTGTCCCTG
TGAACTAGACAAACCTACATTGCAGCCATGAATATATAAGAGGCAGCCAAGATTTACTCATTGTTTACCA
GGGTCTCAAGTTAGCTCCACTCATCTCACCTGTCATCTTTTGAGTCCTCAAGTAATTCACTGCCTTTGTC
CTCACGCCACATCTCCATAAATTAAAATGTTACGTGGGCTCTTCCGGTTTAATAATGGAAAATATTAGTA
CATCAGTCTTAAGTCCTCACATTTTCAGTCATCCATTGAGCAATTAATAGCACTATAGGCCAAATATTAA
ACTCGAAGGGTCAATTGTTGGAGTTTTGGTTATTTGCTTTTTGAATTGCTTACTTGGGACCATTGGAGCT
GCCATTTAAGAACTATCTTAGCATTCTACACTGCAAAAAATGC