ZFLNCG04076
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR630463 | 5 dpf | normal | 88.36 |
| SRR630464 | 5 dpf | injected epidermidis | 80.62 |
| SRR1167762 | embryo | ptpn6 morpholino | 47.86 |
| SRR942769 | embryo | myd88+/+ | 29.30 |
| SRR1167752 | embryo | normal | 17.43 |
| SRR1167763 | embryo | ptpn6 morpholino and bacterial infection | 16.42 |
| SRR1167753 | embryo | bacterial infection | 15.66 |
| SRR535847 | larvae | normal | 12.11 |
| SRR1004786 | larvae | control morpholino | 11.28 |
| SRR535849 | larvae | normal | 8.62 |
Express in tissues
Correlated coding gene
Download
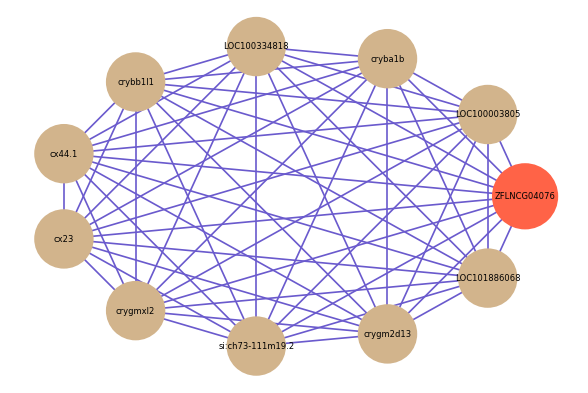
| gene | correlation coefficent |
|---|---|
| LOC100003805 | 0.58 |
| cryba1b | 0.53 |
| LOC100334818 | 0.53 |
| crybb1l1 | 0.53 |
| cx44.1 | 0.53 |
| cx23 | 0.53 |
| crygmxl2 | 0.52 |
| si:ch73-111m19.2 | 0.52 |
| crygm2d13 | 0.52 |
| LOC101886068 | 0.52 |
Gene Ontology
Download
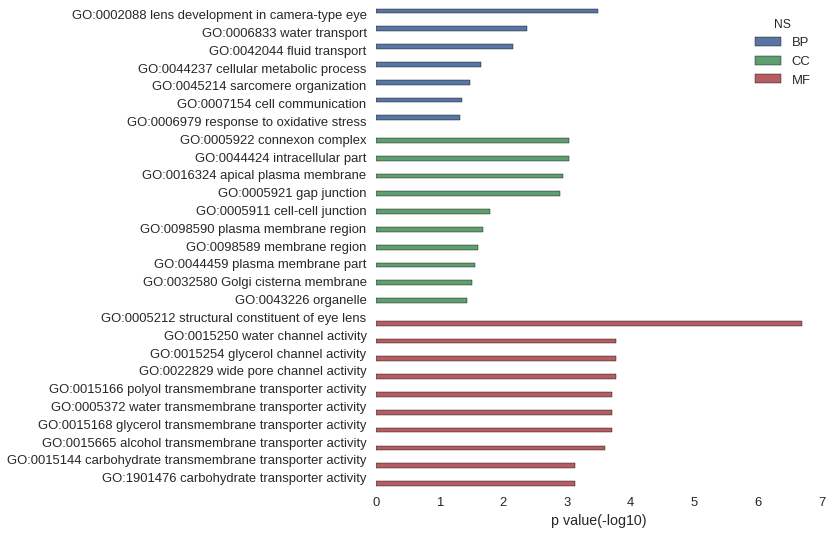
| GO | P value |
|---|---|
| GO:0002088 | 3.23e-04 |
| GO:0006833 | 4.26e-03 |
| GO:0042044 | 7.09e-03 |
| GO:0044237 | 2.25e-02 |
| GO:0045214 | 3.36e-02 |
| GO:0007154 | 4.46e-02 |
| GO:0006979 | 4.73e-02 |
| GO:0005922 | 9.28e-04 |
| GO:0044424 | 9.31e-04 |
| GO:0016324 | 1.17e-03 |
| GO:0005921 | 1.31e-03 |
| GO:0005911 | 1.59e-02 |
| GO:0098590 | 2.08e-02 |
| GO:0098589 | 2.52e-02 |
| GO:0044459 | 2.78e-02 |
| GO:0032580 | 3.08e-02 |
| GO:0043226 | 3.76e-02 |
| GO:0005212 | 2.05e-07 |
| GO:0015250 | 1.73e-04 |
| GO:0015254 | 1.73e-04 |
| GO:0022829 | 1.73e-04 |
| GO:0015166 | 1.99e-04 |
| GO:0005372 | 1.99e-04 |
| GO:0015168 | 1.99e-04 |
| GO:0015665 | 2.58e-04 |
| GO:0015144 | 7.61e-04 |
| GO:1901476 | 7.61e-04 |
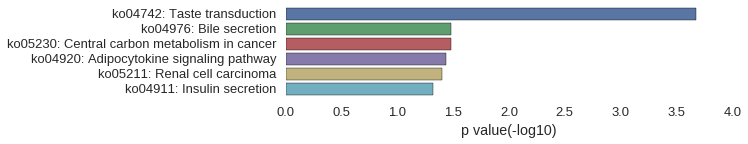
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04076
TCTCTCATCTATTTTTGTTTCATTATACCTAAATATGGTTTGTACGGAAGAACTTCAGTGCTCCAGTCAA
CATCACAGGTATGATGGAACTCACTTATGTTTTGTTTATTATTAACTGGTTGTTAAAAAATGCAATCCTT
TTTGTTTTATTTGTTTCCCCCTCAAAAAGGCATCATGTAAATTATTCATTGTTCAAATGAGCATTCAGAA
AAACAAAATAAAGGTTAGTACATTTTGATGATCCAAAGTGAAAGCTCCAAGTCAGTAAAACGATCAGATC
TTCTCATCACTAAGCGTGTTTTATTACACATACTTGCTTTCTTGTTTCAGATATGAACTGATGCCATCTG
CTGTAGTGAAAAG
TCTCTCATCTATTTTTGTTTCATTATACCTAAATATGGTTTGTACGGAAGAACTTCAGTGCTCCAGTCAA
CATCACAGGTATGATGGAACTCACTTATGTTTTGTTTATTATTAACTGGTTGTTAAAAAATGCAATCCTT
TTTGTTTTATTTGTTTCCCCCTCAAAAAGGCATCATGTAAATTATTCATTGTTCAAATGAGCATTCAGAA
AAACAAAATAAAGGTTAGTACATTTTGATGATCCAAAGTGAAAGCTCCAAGTCAGTAAAACGATCAGATC
TTCTCATCACTAAGCGTGTTTTATTACACATACTTGCTTTCTTGTTTCAGATATGAACTGATGCCATCTG
CTGTAGTGAAAAG