ZFLNCG04077
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 153.70 |
| ERR023143 | swim bladder | normal | 139.18 |
| SRR1562532 | spleen | normal | 73.19 |
| ERR023146 | head kidney | normal | 63.88 |
| SRR1647684 | spleen | SVCV treatment | 62.20 |
| SRR1647682 | spleen | normal | 57.43 |
| SRR592699 | pineal gland | normal | 46.15 |
| SRR592700 | pineal gland | normal | 42.30 |
| SRR1048061 | pineal gland | dark | 38.06 |
| SRR594769 | blood | normal | 31.27 |
Express in tissues
Correlated coding gene
Download
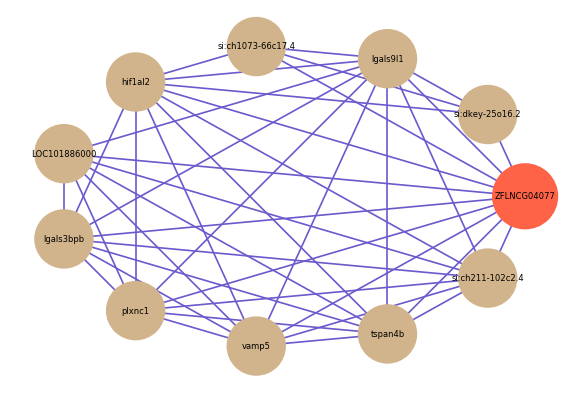
| gene | correlation coefficent |
|---|---|
| si:dkey-25o16.2 | 0.63 |
| LOC101886000 | 0.58 |
| lgals9l1 | 0.57 |
| si:ch1073-66c17.4 | 0.57 |
| hif1al2 | 0.57 |
| lgals3bpb | 0.57 |
| plxnc1 | 0.57 |
| vamp5 | 0.57 |
| tspan4b | 0.56 |
| si:ch211-102c2.4 | 0.56 |
Gene Ontology
Download
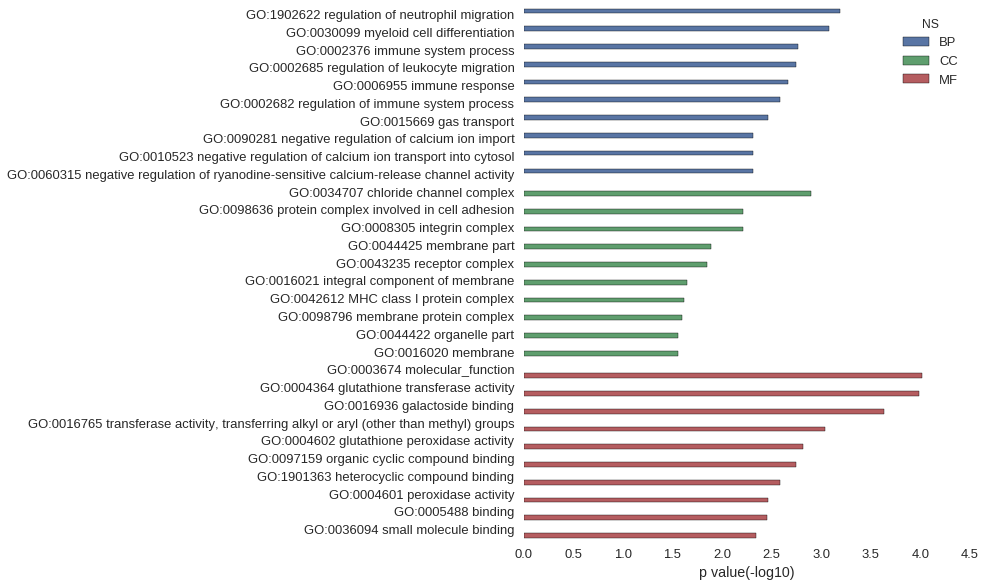
| GO | P value |
|---|---|
| GO:1902622 | 6.51e-04 |
| GO:0030099 | 8.31e-04 |
| GO:0002376 | 1.73e-03 |
| GO:0002685 | 1.78e-03 |
| GO:0006955 | 2.17e-03 |
| GO:0002682 | 2.59e-03 |
| GO:0015669 | 3.45e-03 |
| GO:0090281 | 4.90e-03 |
| GO:0010523 | 4.90e-03 |
| GO:0060315 | 4.90e-03 |
| GO:0034707 | 1.27e-03 |
| GO:0098636 | 6.10e-03 |
| GO:0008305 | 6.10e-03 |
| GO:0044425 | 1.28e-02 |
| GO:0043235 | 1.41e-02 |
| GO:0016021 | 2.23e-02 |
| GO:0042612 | 2.43e-02 |
| GO:0098796 | 2.50e-02 |
| GO:0044422 | 2.77e-02 |
| GO:0016020 | 2.78e-02 |
| GO:0003674 | 9.70e-05 |
| GO:0004364 | 1.03e-04 |
| GO:0016936 | 2.35e-04 |
| GO:0016765 | 9.09e-04 |
| GO:0004602 | 1.52e-03 |
| GO:0097159 | 1.79e-03 |
| GO:1901363 | 2.57e-03 |
| GO:0004601 | 3.45e-03 |
| GO:0005488 | 3.48e-03 |
| GO:0036094 | 4.51e-03 |
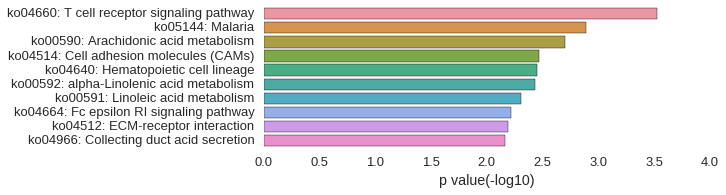
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04077
AATTATTGTATTTACTTTGTCTTATGGTTTGTTTGTTTTTGTTTATTTAAATTTTTCTGGGAGGTGGTTG
TCAGTATAATGATATACTGGTCAGCACCTATAACTAATGCTTAATCCTAAATGCCTTCATTTTGTGTGCA
GTAGATAGTTAATACAACATGTTAACTGCATACAGGATAATCTGCATTCAGCAATTAATTTTACAATAAT
GATATGATTCCAATCCACAGCAATGCATGTTTGATACAGAAGAGTCAGTGAGG
AATTATTGTATTTACTTTGTCTTATGGTTTGTTTGTTTTTGTTTATTTAAATTTTTCTGGGAGGTGGTTG
TCAGTATAATGATATACTGGTCAGCACCTATAACTAATGCTTAATCCTAAATGCCTTCATTTTGTGTGCA
GTAGATAGTTAATACAACATGTTAACTGCATACAGGATAATCTGCATTCAGCAATTAATTTTACAATAAT
GATATGATTCCAATCCACAGCAATGCATGTTTGATACAGAAGAGTCAGTGAGG