ZFLNCG04294
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592703 | pineal gland | normal | 176.22 |
| SRR700534 | heart | control morpholino | 136.29 |
| SRR941753 | posterior pectoral fin | normal | 128.93 |
| SRR1647684 | spleen | SVCV treatment | 127.07 |
| SRR941749 | anterior pectoral fin | normal | 88.85 |
| SRR038626 | embryo | control morpholino and bacterial infection | 83.69 |
| SRR592702 | pineal gland | normal | 77.80 |
| SRR1647683 | spleen | SVCV treatment | 76.80 |
| SRR592699 | pineal gland | normal | 74.65 |
| SRR700537 | heart | Gata4 morpholino | 73.30 |
Express in tissues
Correlated coding gene
Download
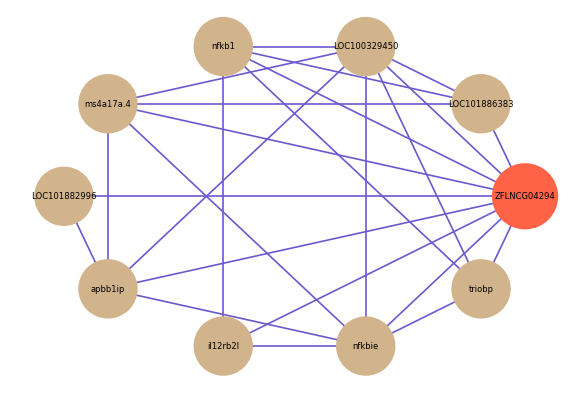
| gene | correlation coefficent |
|---|---|
| LOC101886383 | 0.56 |
| LOC101882996 | 0.55 |
| il12rb2l | 0.53 |
| nfkbie | 0.53 |
| LOC100329450 | 0.53 |
| nfkb1 | 0.52 |
| triobp | 0.51 |
| ms4a17a.4 | 0.51 |
| apbb1ip | 0.51 |
Gene Ontology
Download

| GO | P value |
|---|---|
| GO:0003674 | 2.52e-02 |
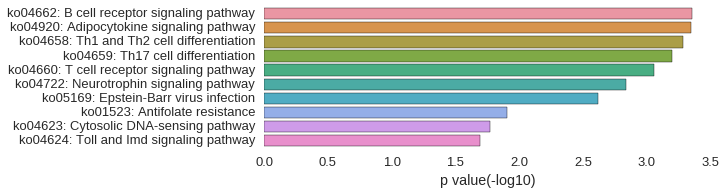
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04294
GCAGATCTTCGCTTCAGCATGTCAGGCGACGCTCGTCACAGTAGGAGGCGACGCCCCCCCAGAATGTATA
TGGTCGCGTCCGACTACAAGGTGCAGGATCACCTGGAAGGAACCATTCGCCTGCAGCGGGGGCAGCTGTG
TGTGGAGCAGGAGGGAGGAGGGGGACACCGCGGAGGAAGAGGAGAACACCTGTGGGTCAAGGTGAGACAC
TGAGAAATACACCTTTACTCACTGTTTACATGCCCACTGTGAGCTTAGATCTCGCTATTTAGAGTTTGTT
TATGTGACGTGAGTCTCTTTTTTTCTCTTTAACATTAAAACAATATTACCTAGTCCTAGGTGACATCTTT
TGATGATGATGTCCGCAGAATGATGAGCTCATACAGTTTTATTTTATTTATAACATAAGATTAGTTTTTT
TTTACCACCGGTAAATGAAGCCTTTTCGATTTTATCTCCAAATGTTTACCATTTCCATCCTAATTGTGAA
GTTTAAACTTTAAAACTTTGGTTGCATATTGCAATTTATACCAATTATGTCCTGAAATAGTCAAACTTCT
GTTACCCCTAGTTCATAAAGAAAAAGGACTTTCACTTAAATAACCTTTATTAATTACACAAAAACACTTT
GGTAGCAGGTGAGCCGAATAATCACAGTTAGGTTTTCATATCCAGTGCAATATTGTCAAGCTAATAGGGA
GTCATACCCCAGTCTTAGATAATATGCAAATCTGCAATACTCTATAGTCTTTATAATAGAAATTCAAAAC
AGTGACATTAACAAACTCCTACACACAGCTACGATAGCTGTTACCCGGCATTAATGACACCCAGCAACAC
CTGCGGCAGGTATCTGCGACTGCACATCCTCAAACACCATTAACGCTGTCCAACCTTCCATAGATGGTCC
ACATTGTCTG
GCAGATCTTCGCTTCAGCATGTCAGGCGACGCTCGTCACAGTAGGAGGCGACGCCCCCCCAGAATGTATA
TGGTCGCGTCCGACTACAAGGTGCAGGATCACCTGGAAGGAACCATTCGCCTGCAGCGGGGGCAGCTGTG
TGTGGAGCAGGAGGGAGGAGGGGGACACCGCGGAGGAAGAGGAGAACACCTGTGGGTCAAGGTGAGACAC
TGAGAAATACACCTTTACTCACTGTTTACATGCCCACTGTGAGCTTAGATCTCGCTATTTAGAGTTTGTT
TATGTGACGTGAGTCTCTTTTTTTCTCTTTAACATTAAAACAATATTACCTAGTCCTAGGTGACATCTTT
TGATGATGATGTCCGCAGAATGATGAGCTCATACAGTTTTATTTTATTTATAACATAAGATTAGTTTTTT
TTTACCACCGGTAAATGAAGCCTTTTCGATTTTATCTCCAAATGTTTACCATTTCCATCCTAATTGTGAA
GTTTAAACTTTAAAACTTTGGTTGCATATTGCAATTTATACCAATTATGTCCTGAAATAGTCAAACTTCT
GTTACCCCTAGTTCATAAAGAAAAAGGACTTTCACTTAAATAACCTTTATTAATTACACAAAAACACTTT
GGTAGCAGGTGAGCCGAATAATCACAGTTAGGTTTTCATATCCAGTGCAATATTGTCAAGCTAATAGGGA
GTCATACCCCAGTCTTAGATAATATGCAAATCTGCAATACTCTATAGTCTTTATAATAGAAATTCAAAAC
AGTGACATTAACAAACTCCTACACACAGCTACGATAGCTGTTACCCGGCATTAATGACACCCAGCAACAC
CTGCGGCAGGTATCTGCGACTGCACATCCTCAAACACCATTAACGCTGTCCAACCTTCCATAGATGGTCC
ACATTGTCTG