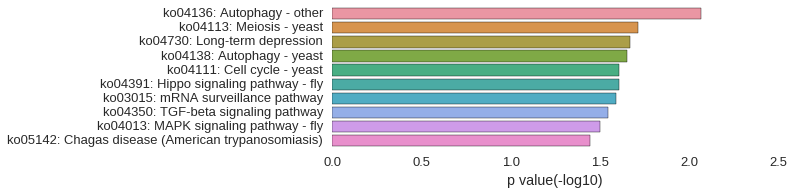
ZFLNCG04295
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 243.25 |
| SRR700534 | heart | control morpholino | 213.79 |
| SRR516121 | skin | male and 3.5 year | 184.04 |
| SRR038626 | embryo | control morpholino and bacterial infection | 149.04 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 142.97 |
| SRR516123 | skin | male and 3.5 year | 139.63 |
| SRR516129 | skin | male and 5 month | 131.22 |
| SRR1291417 | 5 dpi | injected marinum | 127.47 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 109.88 |
| SRR516122 | skin | male and 3.5 year | 108.23 |
Express in tissues
Correlated coding gene
Download
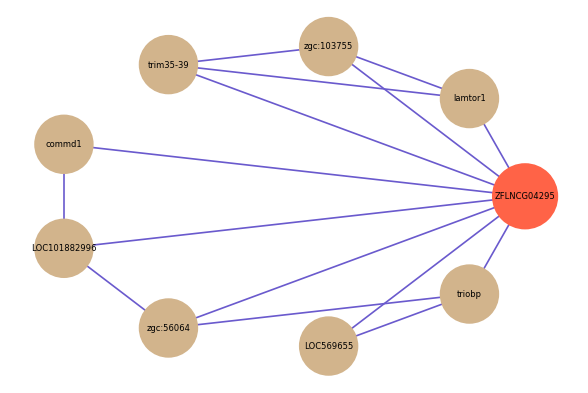
| gene | correlation coefficent |
|---|---|
| lamtor1 | 0.63 |
| commd1 | 0.57 |
| zgc:103755 | 0.53 |
| LOC101882996 | 0.52 |
| LOC569655 | 0.52 |
| trim35-39 | 0.52 |
| triobp | 0.52 |
| zgc:56064 | 0.50 |
Gene Ontology
Download
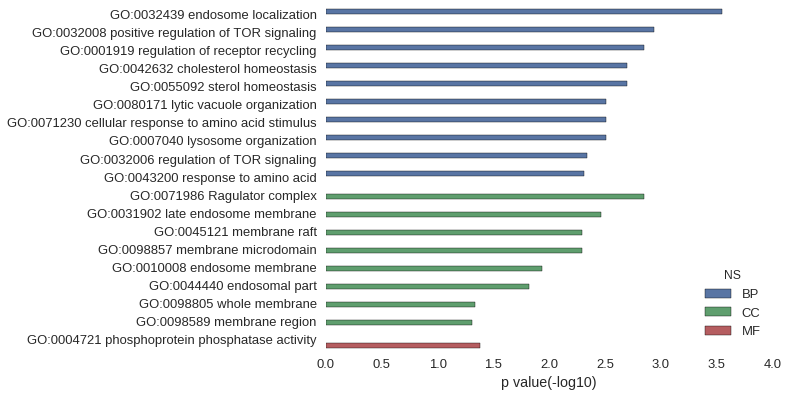
| GO | P value |
|---|---|
| GO:0032439 | 2.84e-04 |
| GO:0032008 | 1.14e-03 |
| GO:0001919 | 1.42e-03 |
| GO:0042632 | 1.99e-03 |
| GO:0055092 | 1.99e-03 |
| GO:0080171 | 3.12e-03 |
| GO:0071230 | 3.12e-03 |
| GO:0007040 | 3.12e-03 |
| GO:0032006 | 4.54e-03 |
| GO:0043200 | 4.82e-03 |
| GO:0071986 | 1.42e-03 |
| GO:0031902 | 3.41e-03 |
| GO:0045121 | 5.11e-03 |
| GO:0098857 | 5.11e-03 |
| GO:0010008 | 1.16e-02 |
| GO:0044440 | 1.50e-02 |
| GO:0098805 | 4.58e-02 |
| GO:0098589 | 4.88e-02 |
| GO:0004721 | 4.17e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04295
ACGGCTTCAACGTCCCGCAAGTGAGCTGGTGTCCGGAGCTGAGCGAGCTCTTCAGTCTGTCTGTGGATGA
AGTGTGTCGTCTGGATGCGCTGCGGGGACCCGTCAGACGCCTGCTCTGTGACTCTTACATGTGTCTGTAC
CACTGCCCGCAGCTCACACTCTACAAATAACACACACACCTCTGCGACTCCTACTTGTGTCTGTATCACT
GCGCACAACTCACACTCTACAAATAACACACTCCCACACACACACACCTGTATATTACACATCTAAA
ACGGCTTCAACGTCCCGCAAGTGAGCTGGTGTCCGGAGCTGAGCGAGCTCTTCAGTCTGTCTGTGGATGA
AGTGTGTCGTCTGGATGCGCTGCGGGGACCCGTCAGACGCCTGCTCTGTGACTCTTACATGTGTCTGTAC
CACTGCCCGCAGCTCACACTCTACAAATAACACACACACCTCTGCGACTCCTACTTGTGTCTGTATCACT
GCGCACAACTCACACTCTACAAATAACACACTCCCACACACACACACCTGTATATTACACATCTAAA