ZFLNCG04337
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 165.48 |
| ERR023143 | swim bladder | normal | 110.62 |
| SRR957180 | heart | 7 days after heart tip amputation | 27.91 |
| ERR023146 | head kidney | normal | 26.50 |
| SRR957181 | heart | normal | 25.21 |
| SRR594769 | blood | normal | 13.92 |
| SRR372793 | shield | normal | 9.92 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 9.73 |
| SRR700534 | heart | control morpholino | 7.68 |
| SRR1035978 | 13 hpf | rx3-/- | 6.84 |
Express in tissues
Correlated coding gene
Download
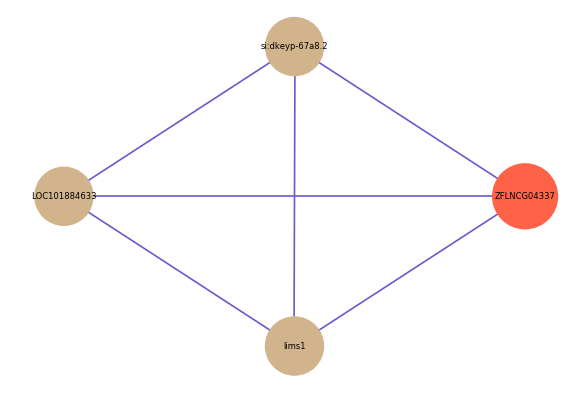
| gene | correlation coefficent |
|---|---|
| si:dkeyp-67a8.2 | 0.60 |
| LOC101884633 | 0.50 |
| lims1 | 0.50 |
Gene Ontology
Download
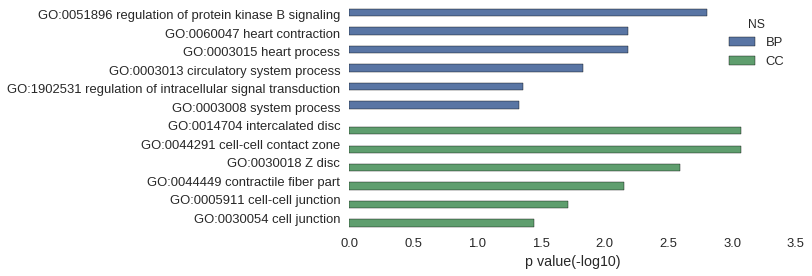
| GO | P value |
|---|---|
| GO:0051896 | 1.56e-03 |
| GO:0060047 | 6.53e-03 |
| GO:0003015 | 6.53e-03 |
| GO:0003013 | 1.47e-02 |
| GO:1902531 | 4.37e-02 |
| GO:0003008 | 4.70e-02 |
| GO:0014704 | 8.53e-04 |
| GO:0044291 | 8.53e-04 |
| GO:0030018 | 2.56e-03 |
| GO:0044449 | 7.09e-03 |
| GO:0005911 | 1.94e-02 |
| GO:0030054 | 3.56e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04337
AAAACGGAAAACTTGATGAGGTTATCAATTCCTTTTTTAATATAAGTAAGGGATAATGTGCACCCAAATG
GTTTTTATCACAGATTAAATCTCAATGTGTTCATCAGCAGCTGTTGTGACTGCAGTTGTTGTAAAAACCG
GCTGGATGTACATTATTCTGTTTATTACACTTGCCACAAAACTAAACAATTGAGACACAAAACATTGATC
TGAGCTGTTTCAATGTACTAGCTCTTTAATAATACATAAAGCCTGCAGAAAGAAAGTTTGTTCTTAACAA
ATGGCTGCAGCCCACGATAACATTGTTATTATTATTTATCGGTCACAAGGTGGCACCAAACAGTCAAGAA
CAGCAGAGTGACAAACGTATAAATTTGTACACGAATATATTCATTAAATGGTTAAGATGAGTCATAGTAC
TTATATTAGTACAATTAGTAGTTTTATCAGTTATTTTCGAAAAATAACACGCCTTAAAATGTTGCCTTTG
ACCAATCAGAATCAAGTATTCCAGAGAGCAGTGTAATAACTGGGAATAACATACCTTTGAATGTTATGAC
TGACCAATCAGAATCAAGTATTCCAGAGAGCAGTTTAATGATGGTAAAAAAAACATACCTTGAAATGTCA
TAACTGACCAATAAGAATCAAGTATTCCAGGGAGCAGTTTAATGATGGTAAAAAAAACATATCTTGAAAT
GTCATAAGAATGAAGTATTCTAGAGAGCCATGTAATAATGGCAAGTAACATAACTTGGAATGTTGAGACA
GACCAATCAGAATCAAGTATTCCAGAGAGCAGTGTAATAACCGGGAATAACATACCTTTGAATGTTGTGA
CTGACCAATCAGAACCAGGTATTCCAGAGAGCCATGTAATAACGAGGAATGAAATATCTTGATTGTTGCA
ACTGA
AAAACGGAAAACTTGATGAGGTTATCAATTCCTTTTTTAATATAAGTAAGGGATAATGTGCACCCAAATG
GTTTTTATCACAGATTAAATCTCAATGTGTTCATCAGCAGCTGTTGTGACTGCAGTTGTTGTAAAAACCG
GCTGGATGTACATTATTCTGTTTATTACACTTGCCACAAAACTAAACAATTGAGACACAAAACATTGATC
TGAGCTGTTTCAATGTACTAGCTCTTTAATAATACATAAAGCCTGCAGAAAGAAAGTTTGTTCTTAACAA
ATGGCTGCAGCCCACGATAACATTGTTATTATTATTTATCGGTCACAAGGTGGCACCAAACAGTCAAGAA
CAGCAGAGTGACAAACGTATAAATTTGTACACGAATATATTCATTAAATGGTTAAGATGAGTCATAGTAC
TTATATTAGTACAATTAGTAGTTTTATCAGTTATTTTCGAAAAATAACACGCCTTAAAATGTTGCCTTTG
ACCAATCAGAATCAAGTATTCCAGAGAGCAGTGTAATAACTGGGAATAACATACCTTTGAATGTTATGAC
TGACCAATCAGAATCAAGTATTCCAGAGAGCAGTTTAATGATGGTAAAAAAAACATACCTTGAAATGTCA
TAACTGACCAATAAGAATCAAGTATTCCAGGGAGCAGTTTAATGATGGTAAAAAAAACATATCTTGAAAT
GTCATAAGAATGAAGTATTCTAGAGAGCCATGTAATAATGGCAAGTAACATAACTTGGAATGTTGAGACA
GACCAATCAGAATCAAGTATTCCAGAGAGCAGTGTAATAACCGGGAATAACATACCTTTGAATGTTGTGA
CTGACCAATCAGAACCAGGTATTCCAGAGAGCCATGTAATAACGAGGAATGAAATATCTTGATTGTTGCA
ACTGA
