ZFLNCG04343
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR801555 | embryo | RPS19 morpholino | 76.25 |
| ERR023144 | brain | normal | 46.96 |
| SRR801556 | embryo | RPS19 and p53 morpholino | 24.57 |
| SRR516122 | skin | male and 3.5 year | 20.52 |
| SRR1708346 | embryo | SIBPS | 20.01 |
| ERR594416 | retina | transgenic flk1 | 18.17 |
| SRR1565817 | brain | normal | 16.96 |
| SRR1005529 | embryo | control morpholino | 14.77 |
| SRR801554 | embryo | control morpholino | 14.74 |
| SRR1565811 | brain | normal | 13.82 |
Express in tissues
Correlated coding gene
Download
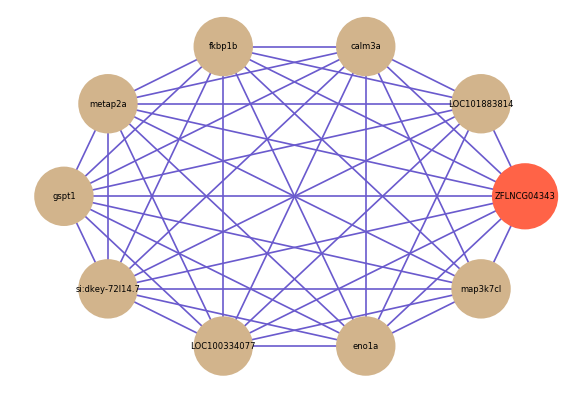
| gene | correlation coefficent |
|---|---|
| LOC101883814 | 0.62 |
| map3k7cl | 0.55 |
| calm3a | 0.54 |
| fkbp1b | 0.53 |
| metap2a | 0.52 |
| gspt1 | 0.52 |
| si:dkey-72l14.7 | 0.51 |
| LOC100334077 | 0.51 |
| eno1a | 0.50 |
Gene Ontology
Download
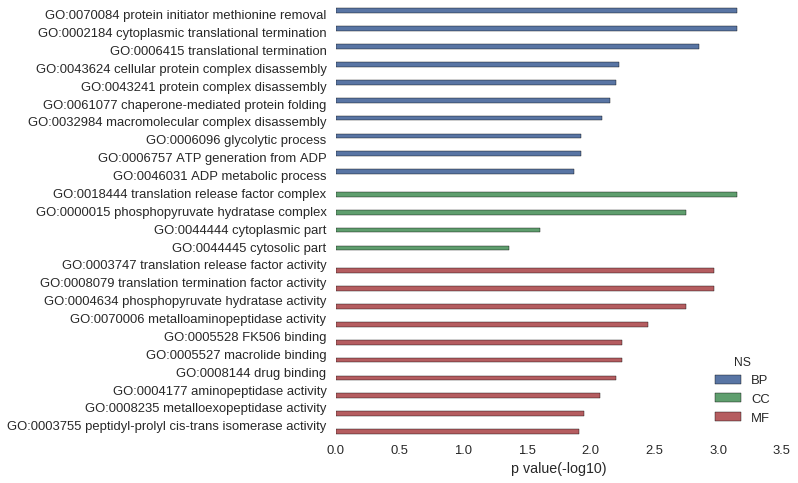
| GO | P value |
|---|---|
| GO:0070084 | 7.11e-04 |
| GO:0002184 | 7.11e-04 |
| GO:0006415 | 1.42e-03 |
| GO:0043624 | 6.03e-03 |
| GO:0043241 | 6.38e-03 |
| GO:0061077 | 7.09e-03 |
| GO:0032984 | 8.15e-03 |
| GO:0006096 | 1.20e-02 |
| GO:0006757 | 1.20e-02 |
| GO:0046031 | 1.34e-02 |
| GO:0018444 | 7.11e-04 |
| GO:0000015 | 1.78e-03 |
| GO:0044444 | 2.48e-02 |
| GO:0044445 | 4.40e-02 |
| GO:0003747 | 1.07e-03 |
| GO:0008079 | 1.07e-03 |
| GO:0004634 | 1.78e-03 |
| GO:0070006 | 3.55e-03 |
| GO:0005528 | 5.67e-03 |
| GO:0005527 | 5.67e-03 |
| GO:0008144 | 6.38e-03 |
| GO:0004177 | 8.50e-03 |
| GO:0008235 | 1.13e-02 |
| GO:0003755 | 1.24e-02 |
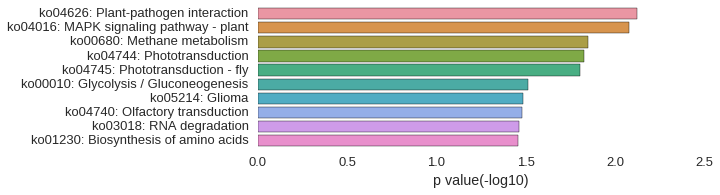
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04343
CCCTTTACTGAGAGAGAAACAGTAGATGATTGTATTGAGACCCATCCTAAAACTCCTTTACACCAGTACT
GACCCTGATCAGACTCAGCAGCTCCTCTGATAGTGAGAGTGTCTCTGTTTACAGTGTAATGACCATCAGA
CTGTAACGTTACACAGTTGCTGTCTTTATACCACTCATATGTCCAGTCACTGTGATCAGACTCAATCACA
CATTTCAGATTGACTGTCTCTCCAGTGAATACAGCACTGTCTGGTGTCACAGTCAATGCTGGTCTTGG
CCCTTTACTGAGAGAGAAACAGTAGATGATTGTATTGAGACCCATCCTAAAACTCCTTTACACCAGTACT
GACCCTGATCAGACTCAGCAGCTCCTCTGATAGTGAGAGTGTCTCTGTTTACAGTGTAATGACCATCAGA
CTGTAACGTTACACAGTTGCTGTCTTTATACCACTCATATGTCCAGTCACTGTGATCAGACTCAATCACA
CATTTCAGATTGACTGTCTCTCCAGTGAATACAGCACTGTCTGGTGTCACAGTCAATGCTGGTCTTGG