ZFLNCG04383
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592698 | pineal gland | normal | 100.70 |
| SRR519747 | 6 dpf | VD3 treatment | 63.14 |
| SRR592699 | pineal gland | normal | 61.14 |
| SRR519727 | 6 dpf | FETOH treatment | 59.16 |
| SRR519752 | 7 dpf | VD3 treatment | 55.48 |
| SRR519722 | 4 dpf | FETOH treatment | 52.68 |
| SRR372802 | 5 dpf | normal | 51.02 |
| SRR519742 | 4 dpf | VD3 treatment | 39.46 |
| SRR519732 | 7 dpf | FETOH treatment | 39.25 |
| SRR1621206 | embryo | normal | 29.79 |
Express in tissues
Correlated coding gene
Download
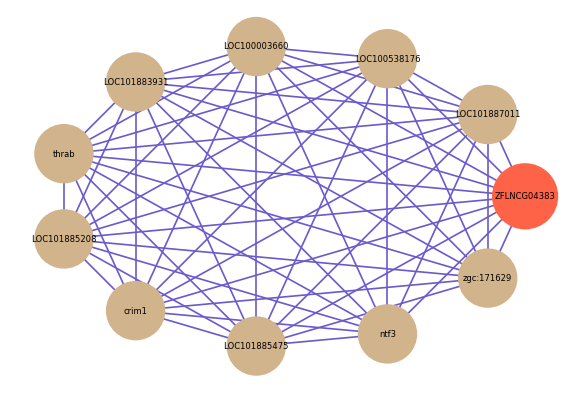
| gene | correlation coefficent |
|---|---|
| LOC101887011 | 0.62 |
| LOC100538176 | 0.61 |
| LOC100003660 | 0.58 |
| LOC101883931 | 0.58 |
| thrab | 0.58 |
| LOC101885208 | 0.58 |
| crim1 | 0.57 |
| LOC101885475 | 0.56 |
| ntf3 | 0.55 |
| zgc:171629 | 0.54 |
Gene Ontology
Download
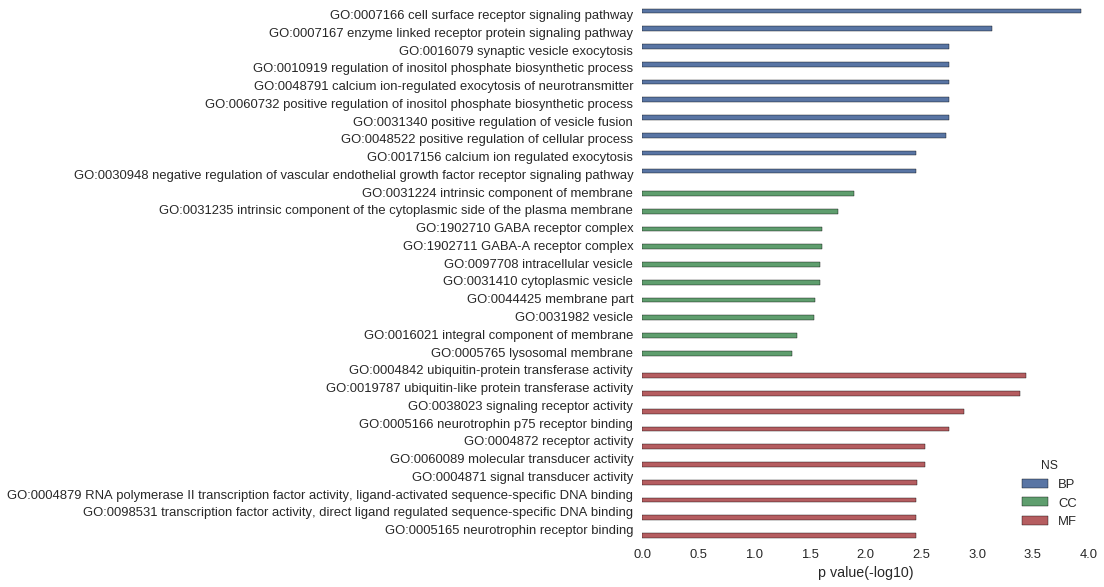
| GO | P value |
|---|---|
| GO:0007166 | 1.17e-04 |
| GO:0007167 | 7.28e-04 |
| GO:0016079 | 1.78e-03 |
| GO:0010919 | 1.78e-03 |
| GO:0048791 | 1.78e-03 |
| GO:0060732 | 1.78e-03 |
| GO:0031340 | 1.78e-03 |
| GO:0048522 | 1.89e-03 |
| GO:0017156 | 3.55e-03 |
| GO:0030948 | 3.55e-03 |
| GO:0031224 | 1.27e-02 |
| GO:0031235 | 1.76e-02 |
| GO:1902710 | 2.46e-02 |
| GO:1902711 | 2.46e-02 |
| GO:0097708 | 2.54e-02 |
| GO:0031410 | 2.54e-02 |
| GO:0044425 | 2.86e-02 |
| GO:0031982 | 2.88e-02 |
| GO:0016021 | 4.14e-02 |
| GO:0005765 | 4.52e-02 |
| GO:0004842 | 3.67e-04 |
| GO:0019787 | 4.12e-04 |
| GO:0038023 | 1.31e-03 |
| GO:0005166 | 1.78e-03 |
| GO:0004872 | 2.95e-03 |
| GO:0060089 | 2.95e-03 |
| GO:0004871 | 3.44e-03 |
| GO:0004879 | 3.52e-03 |
| GO:0098531 | 3.52e-03 |
| GO:0005165 | 3.55e-03 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04383
GATTGCACAGCGGGCGTGTCTTCGGACCGGCGTTGTAAACAGTGTAAAGCAGGAGACTGGAGGTGTGTAT
AAATATCAGTGTGTCGCGCGGGCTGTCTTATGAACTCGTGCCGCGCACCAGCAGCCTGCAGTCCGCACTG
TTTCCGTGGAGCTGACGGCGACCTGCTCACGCCACAGGTTGGTCTGCACTTCAGGAGCTTTATCATTGTG
GATTTCTGATTCATCTTTTGATTTTTGTTTGGAATGATGTCACTGTTGGCTGTACTTGAGTGACTTGCGG
ATTTTGAAGCTCAAGGTGCGTCTCTTCTCATGCGTTGCGCGCTCGCGTGCGTGTTTCGTGGCATTCCTTT
TCTGCCATGTCGCGTTTTAAAGGGCTTTAAAGGCATGAGACTGAGCGGAGATGGCGTTTCCAGTTCTGTG
AGTTTGCGAGTGTTTGTGGCAACATGTTTTTACCGGCGCAAGTTCAGACATAAAAGAGCGATCACGGATT
GTGCCGCGCGCATCCTTTCACCTGAGCGTGCGCGGGCGCGCTATCAGCACCTCATCGATTTCAGTTTCAT
CTGATTAGTACGTTATGAAATAATAACCGTGAGGCGTTTAATGTGTTATGACAACACGTATAATGTTGAA
TAGTCAGTCATGTAAGTGTCGTGTTGTTTAAAGAAGTGTTATGTACTATTTGTTGTACAGTATGGTCGTA
TTTTGAGCTACACTGCCAGTACTATGGTGTTTCCAATAGACAATATTGTGTAAAAAGGACACCTACGCTT
TCATACTGTAACATAATGACACAAAATGCGCTGTAGTAGCTTTTTTTGTAGGTGTATAATATACATAGCG
TCATATTGTATTATCTAACAACGTGTTTTGGTTGTGAAATGTAACGTACTACTTCATTAAGTGTGATTGC
ATTGTACAGTGTGCTGTTGAATGAGGGTTGACAGTGCACTACGCTGCTTAATATTAGTAATATAGCACAT
TATGGGATCTGTTAGGCTACAGTGTAGTTTGAAATGTCATATACTTTCAATGCAAGCACCTGTCGACACC
TGGAAAGTAGGAGC
GATTGCACAGCGGGCGTGTCTTCGGACCGGCGTTGTAAACAGTGTAAAGCAGGAGACTGGAGGTGTGTAT
AAATATCAGTGTGTCGCGCGGGCTGTCTTATGAACTCGTGCCGCGCACCAGCAGCCTGCAGTCCGCACTG
TTTCCGTGGAGCTGACGGCGACCTGCTCACGCCACAGGTTGGTCTGCACTTCAGGAGCTTTATCATTGTG
GATTTCTGATTCATCTTTTGATTTTTGTTTGGAATGATGTCACTGTTGGCTGTACTTGAGTGACTTGCGG
ATTTTGAAGCTCAAGGTGCGTCTCTTCTCATGCGTTGCGCGCTCGCGTGCGTGTTTCGTGGCATTCCTTT
TCTGCCATGTCGCGTTTTAAAGGGCTTTAAAGGCATGAGACTGAGCGGAGATGGCGTTTCCAGTTCTGTG
AGTTTGCGAGTGTTTGTGGCAACATGTTTTTACCGGCGCAAGTTCAGACATAAAAGAGCGATCACGGATT
GTGCCGCGCGCATCCTTTCACCTGAGCGTGCGCGGGCGCGCTATCAGCACCTCATCGATTTCAGTTTCAT
CTGATTAGTACGTTATGAAATAATAACCGTGAGGCGTTTAATGTGTTATGACAACACGTATAATGTTGAA
TAGTCAGTCATGTAAGTGTCGTGTTGTTTAAAGAAGTGTTATGTACTATTTGTTGTACAGTATGGTCGTA
TTTTGAGCTACACTGCCAGTACTATGGTGTTTCCAATAGACAATATTGTGTAAAAAGGACACCTACGCTT
TCATACTGTAACATAATGACACAAAATGCGCTGTAGTAGCTTTTTTTGTAGGTGTATAATATACATAGCG
TCATATTGTATTATCTAACAACGTGTTTTGGTTGTGAAATGTAACGTACTACTTCATTAAGTGTGATTGC
ATTGTACAGTGTGCTGTTGAATGAGGGTTGACAGTGCACTACGCTGCTTAATATTAGTAATATAGCACAT
TATGGGATCTGTTAGGCTACAGTGTAGTTTGAAATGTCATATACTTTCAATGCAAGCACCTGTCGACACC
TGGAAAGTAGGAGC