ZFLNCG04507
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023146 | head kidney | normal | 12.82 |
| SRR1647680 | head kidney | SVCV treatment | 4.76 |
| SRR1647681 | head kidney | SVCV treatment | 2.78 |
| SRR1647679 | head kidney | normal | 2.23 |
| SRR776509 | embryo | Salmonella infected | 1.67 |
| SRR1342217 | embryo | infection with Mycobacterium marinum | 1.57 |
| SRR942772 | embryo | myd88+/+ and infection with Mycobacterium marinum | 1.47 |
| SRR776507 | embryo | normal | 1.44 |
| SRR942769 | embryo | myd88+/+ | 1.40 |
| SRR1167753 | embryo | bacterial infection | 1.29 |
Express in tissues
Correlated coding gene
Download
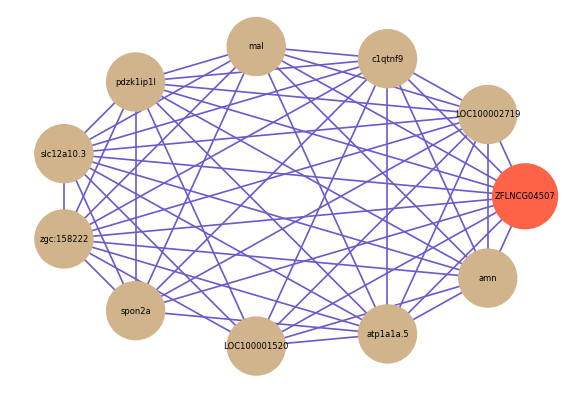
| gene | correlation coefficent |
|---|---|
| LOC100002719 | 0.69 |
| c1qtnf9 | 0.60 |
| mal | 0.59 |
| pdzk1ip1l | 0.58 |
| slc12a10.3 | 0.57 |
| zgc:158222 | 0.56 |
| spon2a | 0.56 |
| LOC100001520 | 0.56 |
| atp1a1a.5 | 0.56 |
| amn | 0.55 |
Gene Ontology
Download
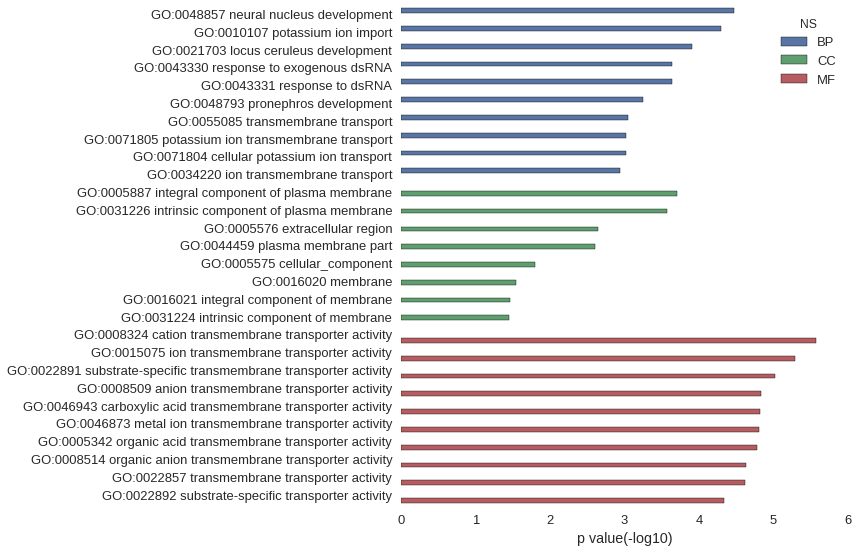
| GO | P value |
|---|---|
| GO:0048857 | 3.40e-05 |
| GO:0010107 | 5.05e-05 |
| GO:0021703 | 1.23e-04 |
| GO:0043330 | 2.29e-04 |
| GO:0043331 | 2.29e-04 |
| GO:0048793 | 5.72e-04 |
| GO:0055085 | 9.09e-04 |
| GO:0071805 | 9.66e-04 |
| GO:0071804 | 9.66e-04 |
| GO:0034220 | 1.14e-03 |
| GO:0005887 | 1.99e-04 |
| GO:0031226 | 2.67e-04 |
| GO:0005576 | 2.29e-03 |
| GO:0044459 | 2.47e-03 |
| GO:0005575 | 1.60e-02 |
| GO:0016020 | 2.89e-02 |
| GO:0016021 | 3.46e-02 |
| GO:0031224 | 3.52e-02 |
| GO:0008324 | 2.69e-06 |
| GO:0015075 | 5.06e-06 |
| GO:0022891 | 9.58e-06 |
| GO:0008509 | 1.46e-05 |
| GO:0046943 | 1.53e-05 |
| GO:0046873 | 1.55e-05 |
| GO:0005342 | 1.67e-05 |
| GO:0008514 | 2.34e-05 |
| GO:0022857 | 2.40e-05 |
| GO:0022892 | 4.57e-05 |
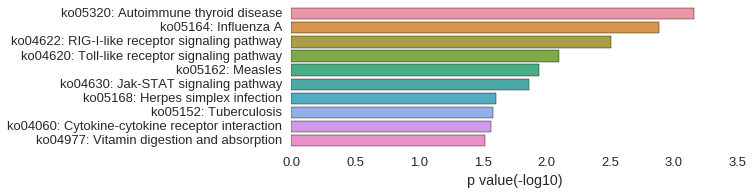
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04507
AGCAGTAGGTTTTTGTGTCTCTGTGTCAGGTTTCTTTGCTGGTGTTTTGTTTTTTCTGAATGCTATCTAA
ATGTAGAAGTAGTGTAACCCTGCCGGTCCAACAGCACCCAACAAGCTGGGATCGAACCAGTGACCTTTTG
CATGAGAGTCGGTTTCTCTAACAAGGAGGCTAAGGACCATGGCCTCTAGCGTATGTCGCTAGAGCAGCTT
TAGAGGAGAGAGGAGTGAAGTTTACCTGAACAGCACTTCAGTAGCAAGCTCAAACTTGCTCGTTTTTGAG
GCGGGAACCGGCGTACGTGCAACAACTTTAATCCTAAGGTAAACACAAAACAAACGTTTCCATCTTGACC
TCTTTCATGGGACTCGACACTAGTTAACATACGCTCCAACGAGTTCCAGTGGCTCTCGGTCCTGCCCATA
CTCATCAATTCTACCAAGCTGACCAATCACAGAGCTTGCGATATGCGTCGCGCTTGACAGAAGTATAAAT
CAGCCTTTAGCCTGGTGTGCATGGCTTCATGCGGGATTCTTTCCAAATAAACTGTGATGTTTGTTTGTTT
GTTTTATTCATTATTATTTGTATAATACTGTAAATTTATTTGAAACATGAAGGATCTTTTAGTTGTTTTG
TCTTTTTTAAAACTCCTTACAGATTGTTTTACATTTTGATTGTTGGGGGGGGGGGTTGATTTGTTATTGA
AGAACTATTGATTAGGTAGAATTTTGAAATTATTTAAAGTATATTAATATATATTGCAAACTCATTTATA
GAGTGTTCTTTTTGCAGTTGTTGGCATACCTCAGAGTGAAGACATTTTGTGCACTGTCGTGGTTTAATAT
GTCATGTTGT
AGCAGTAGGTTTTTGTGTCTCTGTGTCAGGTTTCTTTGCTGGTGTTTTGTTTTTTCTGAATGCTATCTAA
ATGTAGAAGTAGTGTAACCCTGCCGGTCCAACAGCACCCAACAAGCTGGGATCGAACCAGTGACCTTTTG
CATGAGAGTCGGTTTCTCTAACAAGGAGGCTAAGGACCATGGCCTCTAGCGTATGTCGCTAGAGCAGCTT
TAGAGGAGAGAGGAGTGAAGTTTACCTGAACAGCACTTCAGTAGCAAGCTCAAACTTGCTCGTTTTTGAG
GCGGGAACCGGCGTACGTGCAACAACTTTAATCCTAAGGTAAACACAAAACAAACGTTTCCATCTTGACC
TCTTTCATGGGACTCGACACTAGTTAACATACGCTCCAACGAGTTCCAGTGGCTCTCGGTCCTGCCCATA
CTCATCAATTCTACCAAGCTGACCAATCACAGAGCTTGCGATATGCGTCGCGCTTGACAGAAGTATAAAT
CAGCCTTTAGCCTGGTGTGCATGGCTTCATGCGGGATTCTTTCCAAATAAACTGTGATGTTTGTTTGTTT
GTTTTATTCATTATTATTTGTATAATACTGTAAATTTATTTGAAACATGAAGGATCTTTTAGTTGTTTTG
TCTTTTTTAAAACTCCTTACAGATTGTTTTACATTTTGATTGTTGGGGGGGGGGGTTGATTTGTTATTGA
AGAACTATTGATTAGGTAGAATTTTGAAATTATTTAAAGTATATTAATATATATTGCAAACTCATTTATA
GAGTGTTCTTTTTGCAGTTGTTGGCATACCTCAGAGTGAAGACATTTTGTGCACTGTCGTGGTTTAATAT
GTCATGTTGT