ZFLNCG04563
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1048061 | pineal gland | dark | 10.22 |
| SRR726542 | 5 dpf | infection with control | 7.71 |
| SRR592701 | pineal gland | normal | 6.95 |
| SRR516129 | skin | male and 5 month | 6.80 |
| SRR800045 | muscle | normal | 6.76 |
| SRR592703 | pineal gland | normal | 6.59 |
| SRR891511 | brain | normal | 6.41 |
| SRR594769 | blood | normal | 5.64 |
| SRR776508 | embryo | miR-146 knockdown | 5.35 |
| SRR527833 | 5 dpf | normal | 5.13 |
Express in tissues
Correlated coding gene
Download
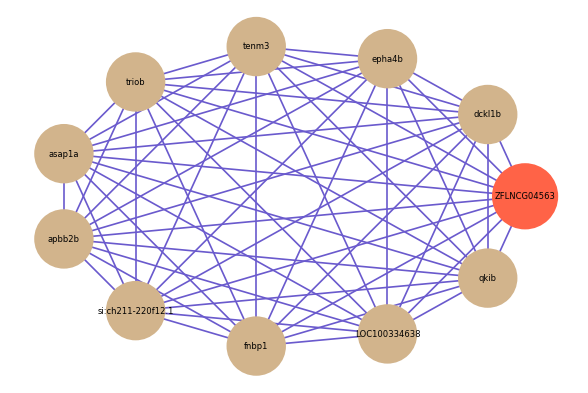
| gene | correlation coefficent |
|---|---|
| dckl1b | 0.59 |
| epha4b | 0.57 |
| tenm3 | 0.55 |
| triob | 0.55 |
| asap1a | 0.55 |
| apbb2b | 0.54 |
| si:ch211-220f12.1 | 0.54 |
| fnbp1 | 0.53 |
| LOC100334638 | 0.53 |
| qkib | 0.53 |
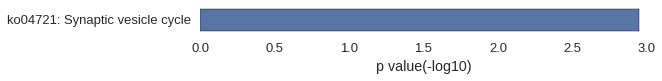
Gene Ontology
Download
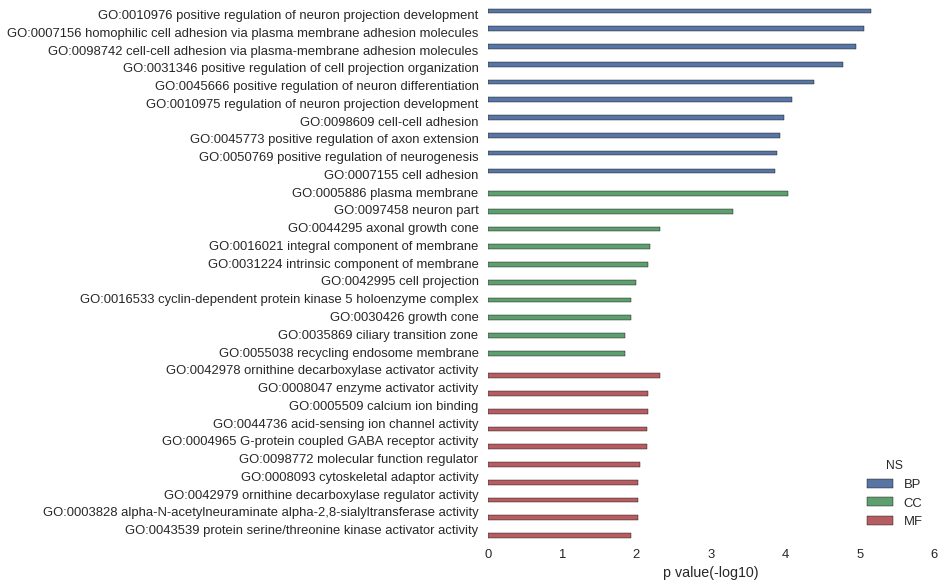
| GO | P value |
|---|---|
| GO:0010976 | 7.06e-06 |
| GO:0007156 | 8.72e-06 |
| GO:0098742 | 1.11e-05 |
| GO:0031346 | 1.66e-05 |
| GO:0045666 | 4.05e-05 |
| GO:0010975 | 8.06e-05 |
| GO:0098609 | 1.05e-04 |
| GO:0045773 | 1.18e-04 |
| GO:0050769 | 1.29e-04 |
| GO:0007155 | 1.38e-04 |
| GO:0005886 | 9.17e-05 |
| GO:0097458 | 5.08e-04 |
| GO:0044295 | 4.83e-03 |
| GO:0016021 | 6.65e-03 |
| GO:0031224 | 6.94e-03 |
| GO:0042995 | 1.02e-02 |
| GO:0016533 | 1.20e-02 |
| GO:0030426 | 1.20e-02 |
| GO:0035869 | 1.44e-02 |
| GO:0055038 | 1.44e-02 |
| GO:0042978 | 4.83e-03 |
| GO:0008047 | 7.02e-03 |
| GO:0005509 | 7.10e-03 |
| GO:0044736 | 7.23e-03 |
| GO:0004965 | 7.23e-03 |
| GO:0098772 | 8.85e-03 |
| GO:0008093 | 9.63e-03 |
| GO:0042979 | 9.63e-03 |
| GO:0003828 | 9.63e-03 |
| GO:0043539 | 1.20e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04563
GAACTACTACTGCAAAGGGAATTTTTCCTTCACAAAGCAGGATATCATCTCTCACTGCCTGACGCAAATG
TACATCCATCCTAAGAACATGTGGAGTTCCTGTCTGTCATTCTAATGAGGGATCTTTAGTGTGCTGGAGG
AACACGCTCTGTGCTCGGACACCATTTGCCGGCCGCTTTCTTTCTGTGTGGATTTACATTAAACGACCAA
GTGTACAGCACAAAGCTGCAGTCTGGATGGATTTGGGTCGAACTAAAATAATAATGTAT
GAACTACTACTGCAAAGGGAATTTTTCCTTCACAAAGCAGGATATCATCTCTCACTGCCTGACGCAAATG
TACATCCATCCTAAGAACATGTGGAGTTCCTGTCTGTCATTCTAATGAGGGATCTTTAGTGTGCTGGAGG
AACACGCTCTGTGCTCGGACACCATTTGCCGGCCGCTTTCTTTCTGTGTGGATTTACATTAAACGACCAA
GTGTACAGCACAAAGCTGCAGTCTGGATGGATTTGGGTCGAACTAAAATAATAATGTAT