ZFLNCG04643
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 40.51 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 36.01 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 35.45 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 33.57 |
| SRR726542 | 5 dpf | infection with control | 10.61 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 10.55 |
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 9.97 |
| SRR527832 | 16-36 hpf | normal | 8.34 |
| SRR527834 | head | normal | 6.58 |
| SRR038625 | embryo | traf6 morpholino | 5.93 |
Express in tissues
Correlated coding gene
Download
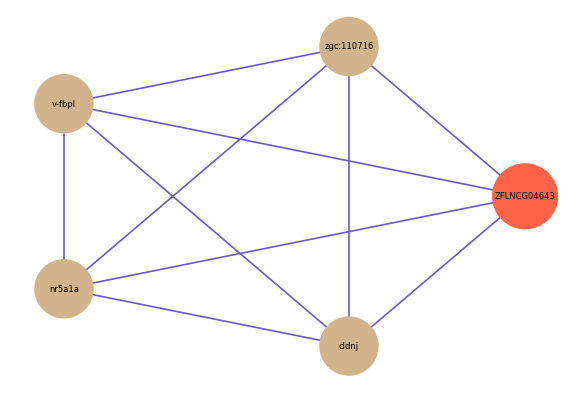
| gene | correlation coefficent |
|---|---|
| zgc:110716 | 0.53 |
| v-fbpl | 0.53 |
| nr5a1a | 0.52 |
| cldnj | 0.51 |
Gene Ontology
Download
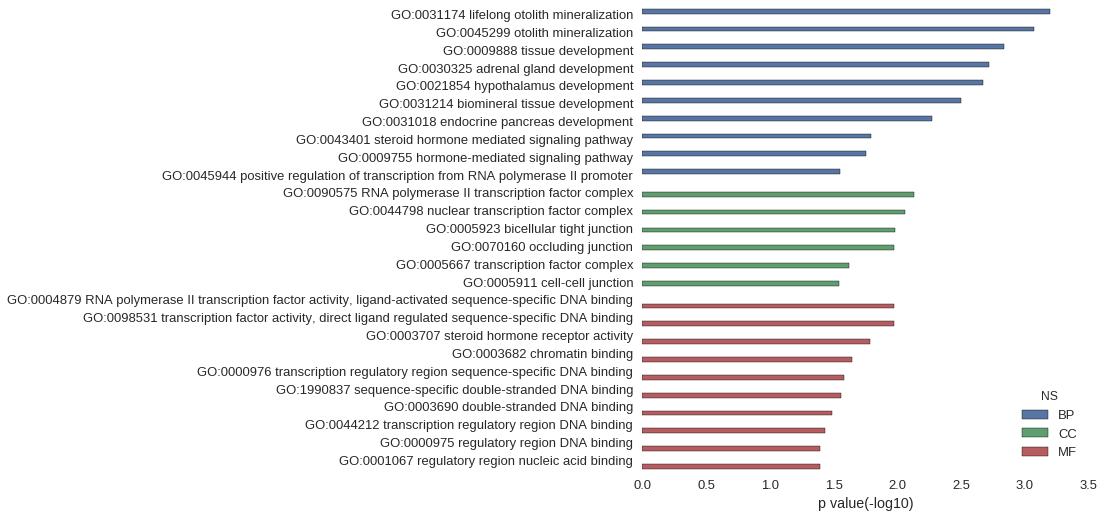
| GO | P value |
|---|---|
| GO:0031174 | 6.39e-04 |
| GO:0045299 | 8.53e-04 |
| GO:0009888 | 1.47e-03 |
| GO:0030325 | 1.92e-03 |
| GO:0021854 | 2.13e-03 |
| GO:0031214 | 3.19e-03 |
| GO:0031018 | 5.32e-03 |
| GO:0043401 | 1.61e-02 |
| GO:0009755 | 1.76e-02 |
| GO:0045944 | 2.81e-02 |
| GO:0090575 | 7.44e-03 |
| GO:0044798 | 8.72e-03 |
| GO:0005923 | 1.04e-02 |
| GO:0070160 | 1.06e-02 |
| GO:0005667 | 2.41e-02 |
| GO:0005911 | 2.89e-02 |
| GO:0004879 | 1.06e-02 |
| GO:0098531 | 1.06e-02 |
| GO:0003707 | 1.65e-02 |
| GO:0003682 | 2.28e-02 |
| GO:0000976 | 2.62e-02 |
| GO:1990837 | 2.75e-02 |
| GO:0003690 | 3.27e-02 |
| GO:0044212 | 3.71e-02 |
| GO:0000975 | 4.04e-02 |
| GO:0001067 | 4.04e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04643
CGTGCCACAGCAGCTGCTGGGGTCTTTAGGTGGGATATATATCTGGTTCTGTTGGATAGATAGCACAGTT
AAGTTAGTAGGTGTTTTGTGCTTGACTTCTGAGTGTCTTTGTTCATGTGTTATACTCACAGGCTGGCAGC
GTGCAGTGCAGTTAGTGCCAGTGGATCTGAGGACGTAGAAACCAGTGGCTGAGTCTAGTGTGTGTGTGCA
CTGTGTGGCGTAACACACACCCTGATCCCGG
CGTGCCACAGCAGCTGCTGGGGTCTTTAGGTGGGATATATATCTGGTTCTGTTGGATAGATAGCACAGTT
AAGTTAGTAGGTGTTTTGTGCTTGACTTCTGAGTGTCTTTGTTCATGTGTTATACTCACAGGCTGGCAGC
GTGCAGTGCAGTTAGTGCCAGTGGATCTGAGGACGTAGAAACCAGTGGCTGAGTCTAGTGTGTGTGTGCA
CTGTGTGGCGTAACACACACCCTGATCCCGG