ZFLNCG04731
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR527832 | 16-36 hpf | normal | 16.24 |
| SRR038626 | embryo | control morpholino and bacterial infection | 13.69 |
| SRR1104059 | heart | kctd10 mut | 12.63 |
| SRR726540 | 5 dpf | normal | 12.51 |
| SRR038625 | embryo | traf6 morpholino | 11.48 |
| SRR038627 | embryo | control morpholino | 9.62 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 8.69 |
| SRR891495 | heart | normal | 8.11 |
| SRR065196 | 3 dpf | normal | 6.80 |
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 6.57 |
Express in tissues
Gene Ontology
Download
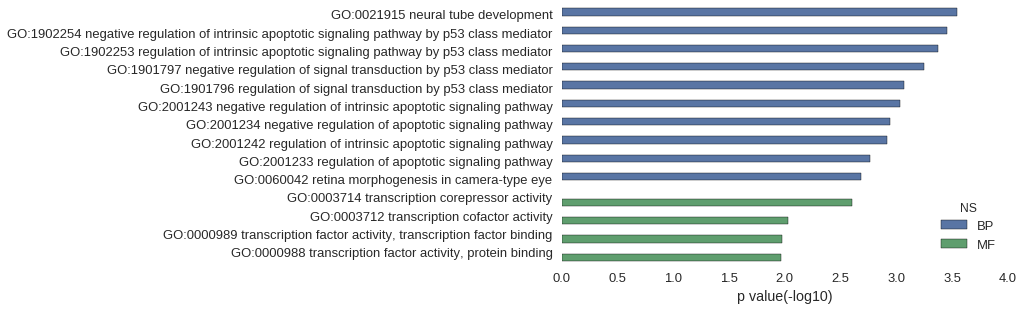
| GO | P value |
|---|---|
| GO:0021915 | 2.84e-04 |
| GO:1902254 | 3.55e-04 |
| GO:1902253 | 4.26e-04 |
| GO:1901797 | 5.69e-04 |
| GO:1901796 | 8.53e-04 |
| GO:2001243 | 9.24e-04 |
| GO:2001234 | 1.14e-03 |
| GO:2001242 | 1.21e-03 |
| GO:2001233 | 1.71e-03 |
| GO:0060042 | 2.06e-03 |
| GO:0003714 | 2.49e-03 |
| GO:0003712 | 9.38e-03 |
| GO:0000989 | 1.07e-02 |
| GO:0000988 | 1.08e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04731
CCCAAAAATGCTTCCGTAATAATCTGATCCCAATCTTTGGAACAATAAAGTACATGCTGACTTAGTCAAG
CACAGATTTGTGTGTGACCATACAACGACTAACAAATCAACATGAAGACTATAAATGTGACGTGTGTGGG
ACAGCATACCCATAACCCCCTCTGTGGTGTTGACAGGCGGGGTGTGAGATGCGGTGACGAAGGGTGAGGT
GCTGGCACTGCTGCGGTGGAAGCTGGACATGGGTGGCAGACTAGCGTTGATGTCTGTGGGCGACACCGAG
TGTGGCGGATAGTTCTGTCACACACAGAGAGAACATGGGAAACTTTTTAGTTTTATAAACAGTTGCACGA
CATGAGAGGTTTGTTTTTGATCTGATCCCGCAGAGCTGGGGATTCGCTGCGTTCTCACCAGGCGGTCGTG
TGAGTGTAGGTTGCTGTAGTTTCCAGACTGTGGCATGTGAGATGAAGATCCTCCCAGCATTCCCCCATAG
CCTGGCTGACTGATCCCATTGGACAAGTTCCACTGGT
CCCAAAAATGCTTCCGTAATAATCTGATCCCAATCTTTGGAACAATAAAGTACATGCTGACTTAGTCAAG
CACAGATTTGTGTGTGACCATACAACGACTAACAAATCAACATGAAGACTATAAATGTGACGTGTGTGGG
ACAGCATACCCATAACCCCCTCTGTGGTGTTGACAGGCGGGGTGTGAGATGCGGTGACGAAGGGTGAGGT
GCTGGCACTGCTGCGGTGGAAGCTGGACATGGGTGGCAGACTAGCGTTGATGTCTGTGGGCGACACCGAG
TGTGGCGGATAGTTCTGTCACACACAGAGAGAACATGGGAAACTTTTTAGTTTTATAAACAGTTGCACGA
CATGAGAGGTTTGTTTTTGATCTGATCCCGCAGAGCTGGGGATTCGCTGCGTTCTCACCAGGCGGTCGTG
TGAGTGTAGGTTGCTGTAGTTTCCAGACTGTGGCATGTGAGATGAAGATCCTCCCAGCATTCCCCCATAG
CCTGGCTGACTGATCCCATTGGACAAGTTCCACTGGT