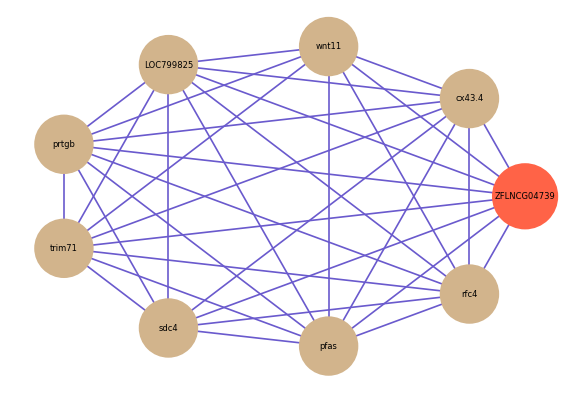
ZFLNCG04739
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR748491 | germ ring | normal | 11.11 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 9.29 |
| SRR372793 | shield | normal | 8.99 |
| ERR023145 | heart | normal | 8.47 |
| SRR1028004 | head | normal | 6.71 |
| ERR023146 | head kidney | normal | 4.12 |
| SRR1004789 | larvae | impdh2 morpholino | 3.92 |
| SRR749515 | 24 hpf | eif3ha morphant | 3.84 |
| SRR1708343 | embryo | MUTAS | 3.36 |
| ERR023143 | swim bladder | normal | 3.15 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
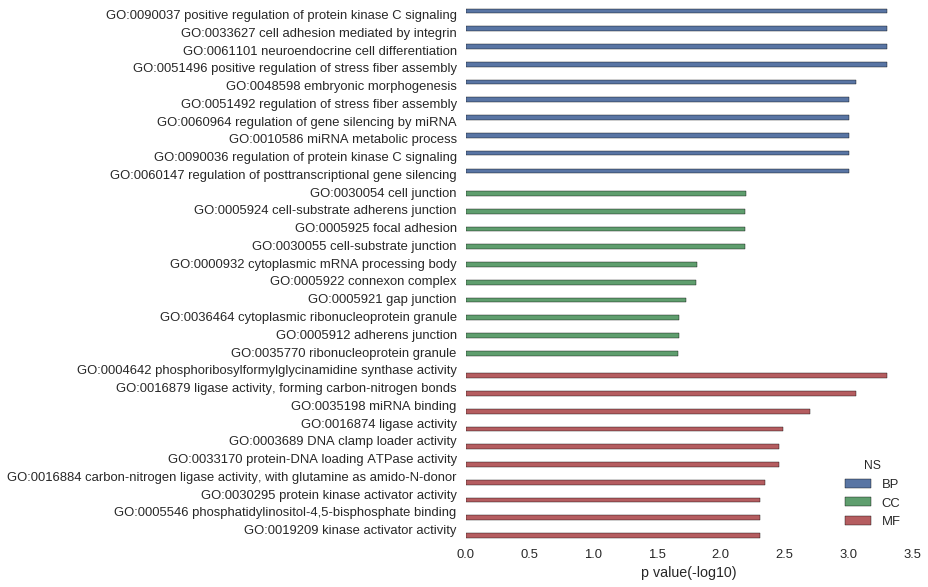
| GO | P value |
|---|---|
| GO:0090037 | 4.97e-04 |
| GO:0033627 | 4.97e-04 |
| GO:0061101 | 4.97e-04 |
| GO:0051496 | 4.97e-04 |
| GO:0048598 | 8.68e-04 |
| GO:0051492 | 9.95e-04 |
| GO:0060964 | 9.95e-04 |
| GO:0010586 | 9.95e-04 |
| GO:0090036 | 9.95e-04 |
| GO:0060147 | 9.95e-04 |
| GO:0030054 | 6.37e-03 |
| GO:0005924 | 6.45e-03 |
| GO:0005925 | 6.45e-03 |
| GO:0030055 | 6.45e-03 |
| GO:0000932 | 1.53e-02 |
| GO:0005922 | 1.58e-02 |
| GO:0005921 | 1.88e-02 |
| GO:0036464 | 2.12e-02 |
| GO:0005912 | 2.12e-02 |
| GO:0035770 | 2.17e-02 |
| GO:0004642 | 4.97e-04 |
| GO:0016879 | 8.69e-04 |
| GO:0035198 | 1.99e-03 |
| GO:0016874 | 3.28e-03 |
| GO:0003689 | 3.48e-03 |
| GO:0033170 | 3.48e-03 |
| GO:0016884 | 4.47e-03 |
| GO:0030295 | 4.96e-03 |
| GO:0005546 | 4.96e-03 |
| GO:0019209 | 4.96e-03 |
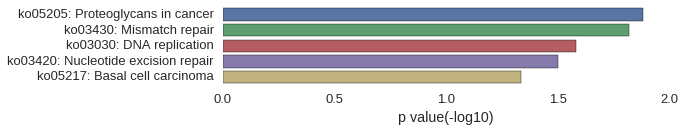
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04739
TATATTTCAAGTTAGGGAGGTTATTGTTGTGGCATTGAGGAGGAAAGGGGCTGATGATGAGGACTCTATC
ACTAATCATGAAGTCAGTCATGCAGGGTCGGAAGAAATACCAGTCAAATGAGTAAACTACTACAGAACTT
GTGCAACATGACTTACTGAAAACTAAGCTGAAATGGTGAGAGATCACAAGAGCAAATCAACTGGATTCAC
TGCAAACCAAATGCATTAGTGGGATGTACAGTAGATGACAAAAACACAAGGTAGGCTATTTCATTGTTTA
TGTCTTGAATGCTAATTTATTTATCTGGATTTTTTTTACTTTTATCATGTGTTTTTCCCTCTATACTCTA
CATTTTAATGTGTTGTTACATTGTATTTTGACCAATAAGAAGTAAATGATTTTGGAAAACTAAACTTACC
AGCCGATGTGTAATCTCTCATTTCAATTTAATTTGCAGACAGAGAGCGGGAGGGGATTTATC
TATATTTCAAGTTAGGGAGGTTATTGTTGTGGCATTGAGGAGGAAAGGGGCTGATGATGAGGACTCTATC
ACTAATCATGAAGTCAGTCATGCAGGGTCGGAAGAAATACCAGTCAAATGAGTAAACTACTACAGAACTT
GTGCAACATGACTTACTGAAAACTAAGCTGAAATGGTGAGAGATCACAAGAGCAAATCAACTGGATTCAC
TGCAAACCAAATGCATTAGTGGGATGTACAGTAGATGACAAAAACACAAGGTAGGCTATTTCATTGTTTA
TGTCTTGAATGCTAATTTATTTATCTGGATTTTTTTTACTTTTATCATGTGTTTTTCCCTCTATACTCTA
CATTTTAATGTGTTGTTACATTGTATTTTGACCAATAAGAAGTAAATGATTTTGGAAAACTAAACTTACC
AGCCGATGTGTAATCTCTCATTTCAATTTAATTTGCAGACAGAGAGCGGGAGGGGATTTATC