ZFLNCG04758
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 74.96 |
| SRR957181 | heart | normal | 45.99 |
| SRR957180 | heart | 7 days after heart tip amputation | 28.38 |
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 26.12 |
| SRR592703 | pineal gland | normal | 24.17 |
| SRR891495 | heart | normal | 24.13 |
| SRR592701 | pineal gland | normal | 20.91 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 16.34 |
| SRR1562531 | muscle | normal | 14.52 |
| ERR023144 | brain | normal | 10.99 |
Express in tissues
Correlated coding gene
Download
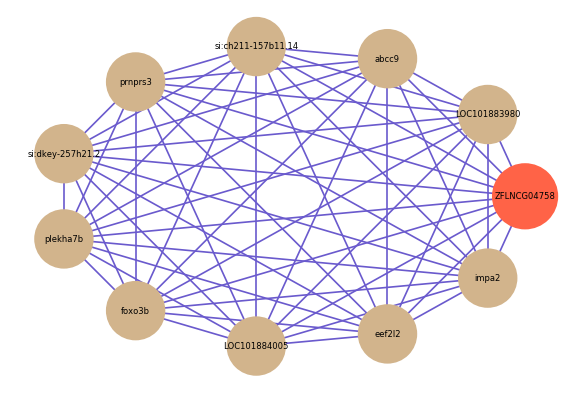
| gene | correlation coefficent |
|---|---|
| LOC101883980 | 0.65 |
| abcc9 | 0.64 |
| si:ch211-157b11.14 | 0.63 |
| prnprs3 | 0.63 |
| si:dkey-257h21.2 | 0.63 |
| plekha7b | 0.63 |
| foxo3b | 0.62 |
| LOC101884005 | 0.62 |
| eef2l2 | 0.62 |
| impa2 | 0.62 |
Gene Ontology
Download
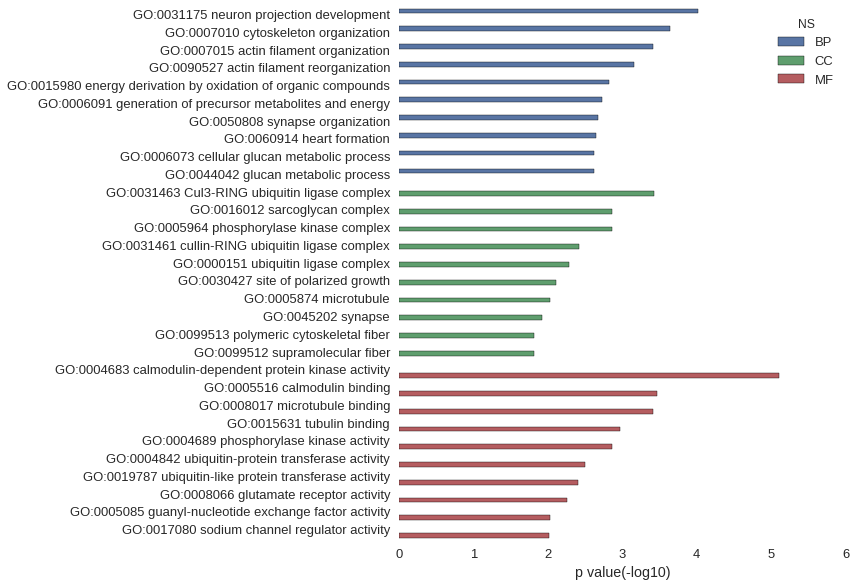
| GO | P value |
|---|---|
| GO:0031175 | 9.59e-05 |
| GO:0007010 | 2.30e-04 |
| GO:0007015 | 3.92e-04 |
| GO:0090527 | 6.90e-04 |
| GO:0015980 | 1.53e-03 |
| GO:0006091 | 1.90e-03 |
| GO:0050808 | 2.13e-03 |
| GO:0060914 | 2.25e-03 |
| GO:0006073 | 2.42e-03 |
| GO:0044042 | 2.42e-03 |
| GO:0031463 | 3.81e-04 |
| GO:0016012 | 1.37e-03 |
| GO:0005964 | 1.37e-03 |
| GO:0031461 | 3.80e-03 |
| GO:0000151 | 5.20e-03 |
| GO:0030427 | 7.79e-03 |
| GO:0005874 | 9.48e-03 |
| GO:0045202 | 1.19e-02 |
| GO:0099513 | 1.52e-02 |
| GO:0099512 | 1.52e-02 |
| GO:0004683 | 7.76e-06 |
| GO:0005516 | 3.38e-04 |
| GO:0008017 | 3.92e-04 |
| GO:0015631 | 1.07e-03 |
| GO:0004689 | 1.37e-03 |
| GO:0004842 | 3.20e-03 |
| GO:0019787 | 3.91e-03 |
| GO:0008066 | 5.62e-03 |
| GO:0005085 | 9.48e-03 |
| GO:0017080 | 9.64e-03 |
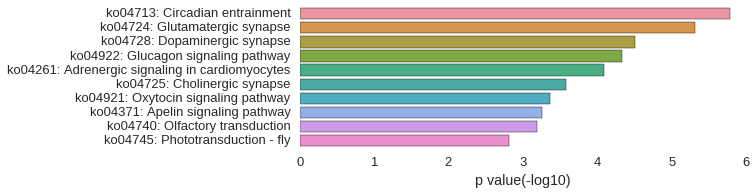
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04758
CACCTTGTGTATATGCAATCTTGATATGTTTGGTCTTGGTGGCATACTGTAGATCTGCTACTGATTAAAT
TATTGGTGCAAAGCAAGTATTGAGACTTGCTACAGTCAGTATATTCACATTGTCAGAAAAAACAGTTTAA
CACTCTGCCATTGCAAACATAACATACAGTAAATATAACCGTCATTGCAGATCTAAGCATAGGTGGAGTT
GGGGCTGAAGAGGGACAGAAAGTTGCAGATTGCTAGTGCAAATGACACTGAGGTACTCTAATGTTGAAGA
GCAAGCTCATTT
CACCTTGTGTATATGCAATCTTGATATGTTTGGTCTTGGTGGCATACTGTAGATCTGCTACTGATTAAAT
TATTGGTGCAAAGCAAGTATTGAGACTTGCTACAGTCAGTATATTCACATTGTCAGAAAAAACAGTTTAA
CACTCTGCCATTGCAAACATAACATACAGTAAATATAACCGTCATTGCAGATCTAAGCATAGGTGGAGTT
GGGGCTGAAGAGGGACAGAAAGTTGCAGATTGCTAGTGCAAATGACACTGAGGTACTCTAATGTTGAAGA
GCAAGCTCATTT