ZFLNCG04841
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1049814 | 256 cell | normal | 18.05 |
| SRR1621206 | embryo | normal | 18.00 |
| SRR372793 | shield | normal | 13.63 |
| SRR372791 | dome | normal | 9.59 |
| SRR372802 | 5 dpf | normal | 9.58 |
| SRR372787 | 2-4 cell | normal | 9.28 |
| SRR700534 | heart | control morpholino | 7.83 |
| SRR891504 | liver | normal | 7.62 |
| SRR372789 | 1K cell | normal | 7.26 |
| SRR658537 | bud | Gata6 morphant | 6.91 |
Express in tissues
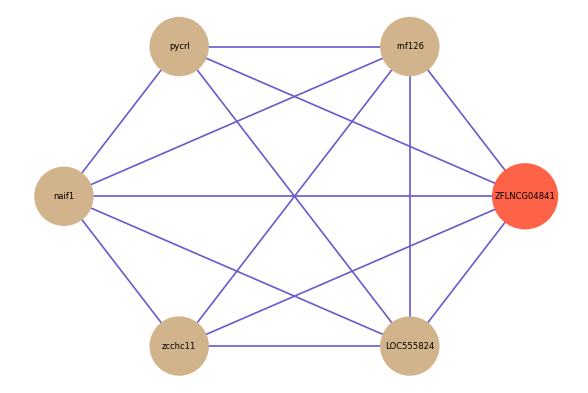
Correlated coding gene
Download
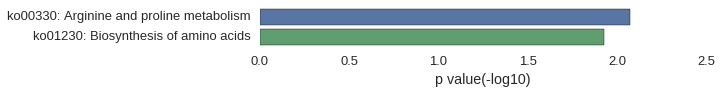
Gene Ontology
Download
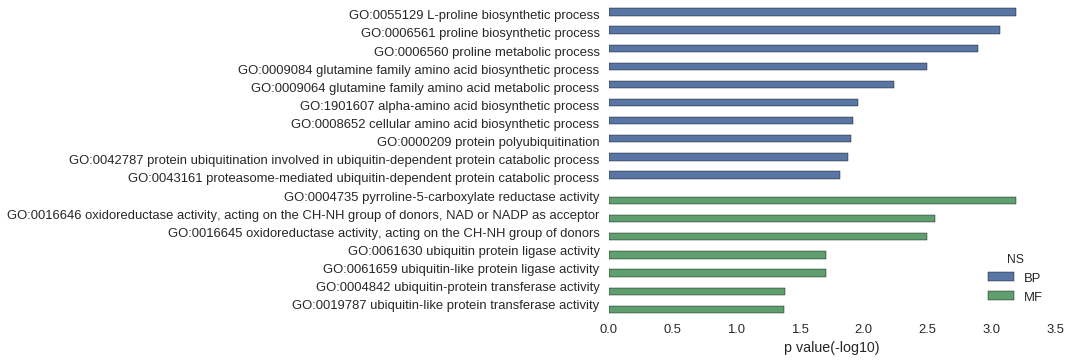
| GO | P value |
|---|---|
| GO:0055129 | 6.39e-04 |
| GO:0006561 | 8.53e-04 |
| GO:0006560 | 1.28e-03 |
| GO:0009084 | 3.19e-03 |
| GO:0009064 | 5.75e-03 |
| GO:1901607 | 1.10e-02 |
| GO:0008652 | 1.21e-02 |
| GO:0000209 | 1.27e-02 |
| GO:0042787 | 1.32e-02 |
| GO:0043161 | 1.53e-02 |
| GO:0004735 | 6.39e-04 |
| GO:0016646 | 2.77e-03 |
| GO:0016645 | 3.19e-03 |
| GO:0061630 | 1.99e-02 |
| GO:0061659 | 1.99e-02 |
| GO:0004842 | 4.12e-02 |
| GO:0019787 | 4.25e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04841
CTCTGTTATTCCTTCGTAACGTCGGTGAATGAACTGCTGCGTGTGTCCGTTGCTGCTAACTGACTGGAGT
GAAGTCAGGCCTCGAATTGAGCGTCGATTTGGCGGAGGATTAAACACATATCTTTAATTTGAGCCACCTT
AAACGGTCATAAACCACAGGTACAACTTGAATTGCCTACCCGGTAGCGTCACGTCGCCGGTGGAAATACA
ACGCGAGGGATAGTTTACCGTCCGTTAGCGGCCGTGCGGATGACAGCGTCCCCGGTGCACGCGCAGAGCG
CCGCCGCGCGCGAGAGCGAGAGCCGCGTTAGCATTAGCCTGTGCGCTTTACTTCCCCTGCATCCCACAGT
CAAACCCAGCAAACGCGAATAAAACCAAGCGGCTTGATCGCTCATAAAAGAAGTCGTTTGACTGCTTGAA
ATAATGTCTCACTCACCAGCCGTTATGATACCAGAAAACAAGTGCAATCTTCTCAATCGACGGAAGGAGG
AACGGACCAGACATTCAAAATGGCGGCGGTCCCAGCCTCAGCACTTCTACCGCAGGCGGAAGGTTACACT
GACTGATGTTCCGAACTTCAAATGCGGATGTGAGAAGGTGCGACCTGCTCTGGTAACACATGATGGGTTT
ATGATGAGCGCTGTCAACATTTATTAGCGCGCTCAAACATTTACAGAAATGAAAGAATGCGGTCGAATGG
CTTGCGGATTTATTAGGTAGCAGAACGCGGTGTATGTCTCCCCATTTAAATATCTCAAGCTTGGTTCTCC
TCGGATATTGTGCGGGTTTTATACTCGTGTTTGTGTCAGTGTAGCACACTTGTGTTTTAAAGCTGGGCTA
CGTGTCAGCTCGGCGGAGGACTTGCCCAAAGAGCCGTGCTGCGGTCCGCACCTCACCTCGTGGCAGTCAC
TGATGACGTCAGCG
CTCTGTTATTCCTTCGTAACGTCGGTGAATGAACTGCTGCGTGTGTCCGTTGCTGCTAACTGACTGGAGT
GAAGTCAGGCCTCGAATTGAGCGTCGATTTGGCGGAGGATTAAACACATATCTTTAATTTGAGCCACCTT
AAACGGTCATAAACCACAGGTACAACTTGAATTGCCTACCCGGTAGCGTCACGTCGCCGGTGGAAATACA
ACGCGAGGGATAGTTTACCGTCCGTTAGCGGCCGTGCGGATGACAGCGTCCCCGGTGCACGCGCAGAGCG
CCGCCGCGCGCGAGAGCGAGAGCCGCGTTAGCATTAGCCTGTGCGCTTTACTTCCCCTGCATCCCACAGT
CAAACCCAGCAAACGCGAATAAAACCAAGCGGCTTGATCGCTCATAAAAGAAGTCGTTTGACTGCTTGAA
ATAATGTCTCACTCACCAGCCGTTATGATACCAGAAAACAAGTGCAATCTTCTCAATCGACGGAAGGAGG
AACGGACCAGACATTCAAAATGGCGGCGGTCCCAGCCTCAGCACTTCTACCGCAGGCGGAAGGTTACACT
GACTGATGTTCCGAACTTCAAATGCGGATGTGAGAAGGTGCGACCTGCTCTGGTAACACATGATGGGTTT
ATGATGAGCGCTGTCAACATTTATTAGCGCGCTCAAACATTTACAGAAATGAAAGAATGCGGTCGAATGG
CTTGCGGATTTATTAGGTAGCAGAACGCGGTGTATGTCTCCCCATTTAAATATCTCAAGCTTGGTTCTCC
TCGGATATTGTGCGGGTTTTATACTCGTGTTTGTGTCAGTGTAGCACACTTGTGTTTTAAAGCTGGGCTA
CGTGTCAGCTCGGCGGAGGACTTGCCCAAAGAGCCGTGCTGCGGTCCGCACCTCACCTCGTGGCAGTCAC
TGATGACGTCAGCG