ZFLNCG04904
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 138.19 |
| SRR592703 | pineal gland | normal | 17.82 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 14.65 |
| SRR1648854 | brain | normal | 14.36 |
| SRR372802 | 5 dpf | normal | 13.82 |
| SRR1035978 | 13 hpf | rx3-/- | 12.93 |
| SRR592701 | pineal gland | normal | 10.02 |
| SRR594771 | endothelium | normal | 8.61 |
| SRR630463 | 5 dpf | normal | 7.65 |
| SRR372793 | shield | normal | 7.52 |
Express in tissues
Correlated coding gene
Download
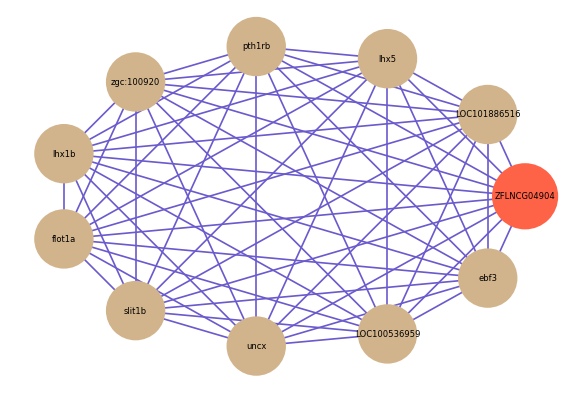
| gene | correlation coefficent |
|---|---|
| LOC101886516 | 0.62 |
| lhx5 | 0.57 |
| pth1rb | 0.57 |
| zgc:100920 | 0.57 |
| lhx1b | 0.56 |
| flot1a | 0.55 |
| slit1b | 0.55 |
| uncx | 0.55 |
| LOC100536959 | 0.54 |
| ebf3 | 0.54 |
Gene Ontology
Download
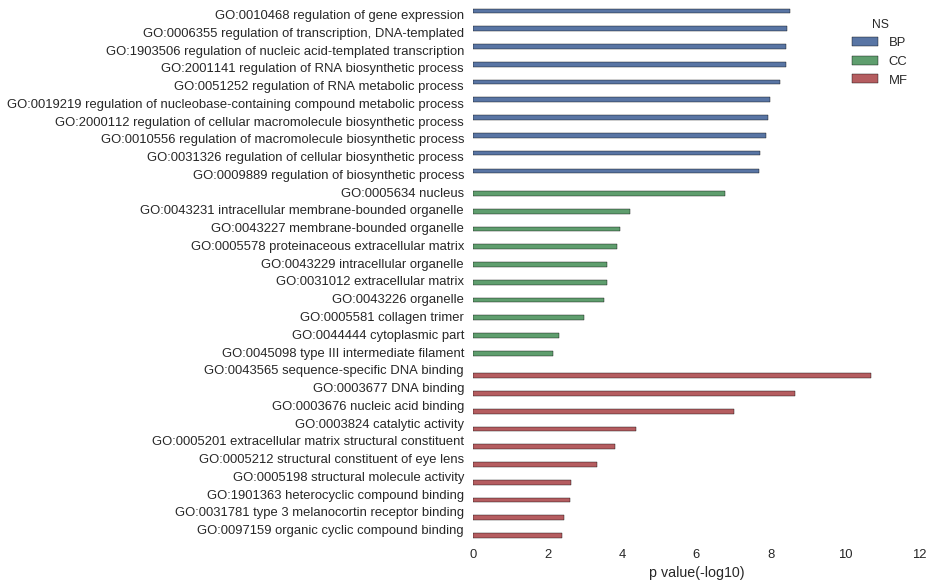
| GO | P value |
|---|---|
| GO:0010468 | 2.99e-09 |
| GO:0006355 | 3.74e-09 |
| GO:1903506 | 3.79e-09 |
| GO:2001141 | 3.89e-09 |
| GO:0051252 | 5.51e-09 |
| GO:0019219 | 1.02e-08 |
| GO:2000112 | 1.16e-08 |
| GO:0010556 | 1.31e-08 |
| GO:0031326 | 1.92e-08 |
| GO:0009889 | 2.03e-08 |
| GO:0005634 | 1.67e-07 |
| GO:0043231 | 6.19e-05 |
| GO:0043227 | 1.12e-04 |
| GO:0005578 | 1.33e-04 |
| GO:0043229 | 2.59e-04 |
| GO:0031012 | 2.59e-04 |
| GO:0043226 | 3.12e-04 |
| GO:0005581 | 1.03e-03 |
| GO:0044444 | 4.87e-03 |
| GO:0045098 | 7.24e-03 |
| GO:0043565 | 1.97e-11 |
| GO:0003677 | 2.26e-09 |
| GO:0003676 | 9.88e-08 |
| GO:0003824 | 4.15e-05 |
| GO:0005201 | 1.53e-04 |
| GO:0005212 | 4.56e-04 |
| GO:0005198 | 2.41e-03 |
| GO:1901363 | 2.49e-03 |
| GO:0031781 | 3.62e-03 |
| GO:0097159 | 4.07e-03 |
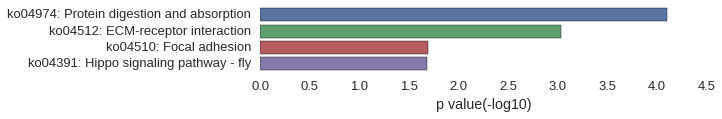
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04904
GACACTGGGTTTCATCCACTAATAATTGCATATGTTTTTCATATTTATATGCACATTTGAACCTCACAAA
AACTGTCCGCCATCTTCACAACACTTCTTCTGTTAGTGAATTTGATTAGTTCTGAGGGGGTGTCCTCGTG
CAGTAATTTGCATAAGACACGCCCACAGCAGCTCCATAGACAGTGTTCAGCATATATAAGTACACGCCTC
ACAAATCTCTCTTTAAATTAATATTTT
GACACTGGGTTTCATCCACTAATAATTGCATATGTTTTTCATATTTATATGCACATTTGAACCTCACAAA
AACTGTCCGCCATCTTCACAACACTTCTTCTGTTAGTGAATTTGATTAGTTCTGAGGGGGTGTCCTCGTG
CAGTAATTTGCATAAGACACGCCCACAGCAGCTCCATAGACAGTGTTCAGCATATATAAGTACACGCCTC
ACAAATCTCTCTTTAAATTAATATTTT