ZFLNCG04929
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR891495 | heart | normal | 9.66 |
| SRR516125 | skin | male and 3.5 year | 8.70 |
| SRR891504 | liver | normal | 6.59 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 6.54 |
| SRR516124 | skin | male and 3.5 year | 6.33 |
| SRR516135 | skin | male and 5 month | 5.62 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 4.88 |
| SRR1534350 | larvae | 5 ug/l BDE47 treatment | 4.82 |
| SRR1028002 | head | normal | 3.34 |
| SRR516130 | skin | male and 5 month | 3.15 |
Express in tissues
Correlated coding gene
Download
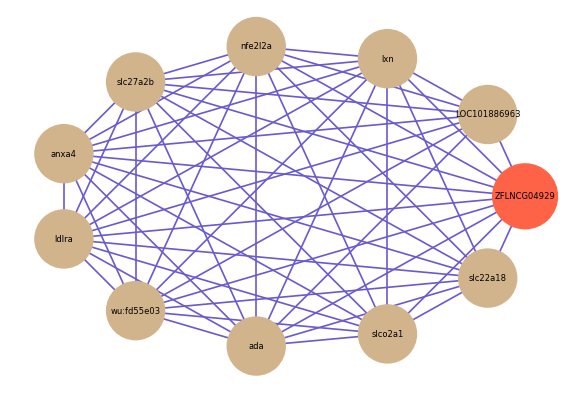
| gene | correlation coefficent |
|---|---|
| LOC101886963 | 0.64 |
| lxn | 0.62 |
| nfe2l2a | 0.62 |
| slc27a2b | 0.61 |
| anxa4 | 0.61 |
| ldlra | 0.60 |
| slco2a1 | 0.60 |
| slc22a18 | 0.59 |
| wu:fd55e03 | 0.58 |
| ada | 0.58 |
Gene Ontology
Download
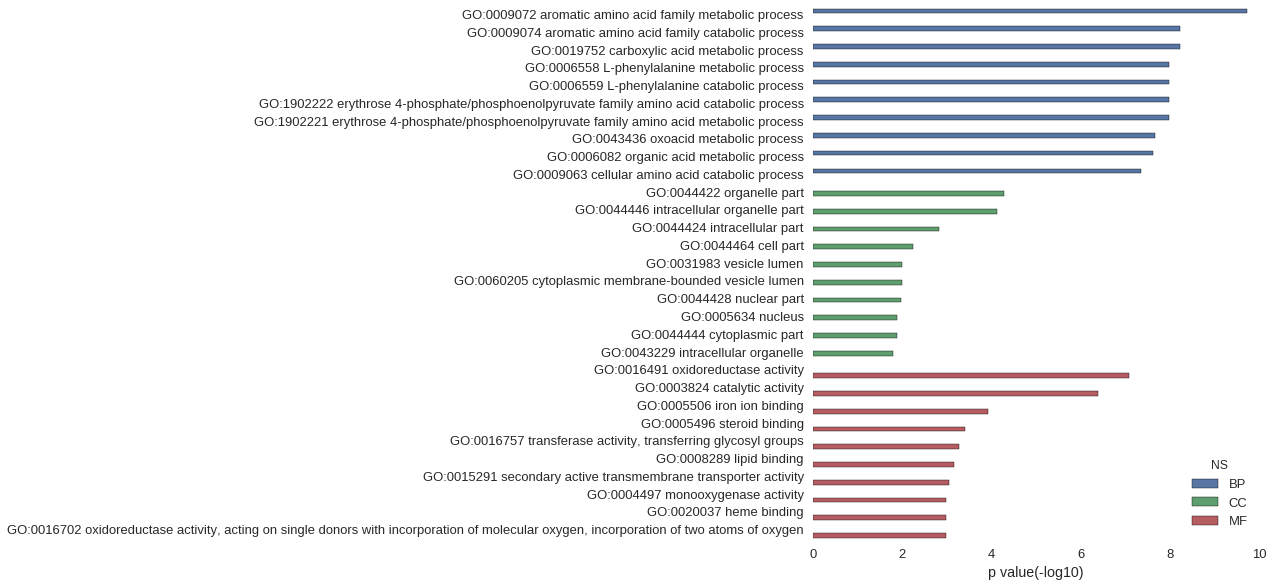
| GO | P value |
|---|---|
| GO:0009072 | 1.90e-10 |
| GO:0009074 | 5.92e-09 |
| GO:0019752 | 5.95e-09 |
| GO:0006558 | 1.08e-08 |
| GO:0006559 | 1.08e-08 |
| GO:1902222 | 1.08e-08 |
| GO:1902221 | 1.08e-08 |
| GO:0043436 | 2.15e-08 |
| GO:0006082 | 2.43e-08 |
| GO:0009063 | 4.55e-08 |
| GO:0044422 | 5.21e-05 |
| GO:0044446 | 7.58e-05 |
| GO:0044424 | 1.47e-03 |
| GO:0044464 | 5.78e-03 |
| GO:0031983 | 1.03e-02 |
| GO:0060205 | 1.03e-02 |
| GO:0044428 | 1.09e-02 |
| GO:0005634 | 1.29e-02 |
| GO:0044444 | 1.29e-02 |
| GO:0043229 | 1.59e-02 |
| GO:0016491 | 8.20e-08 |
| GO:0003824 | 4.21e-07 |
| GO:0005506 | 1.21e-04 |
| GO:0005496 | 3.94e-04 |
| GO:0016757 | 5.30e-04 |
| GO:0008289 | 6.91e-04 |
| GO:0015291 | 8.85e-04 |
| GO:0004497 | 1.05e-03 |
| GO:0020037 | 1.05e-03 |
| GO:0016702 | 1.07e-03 |
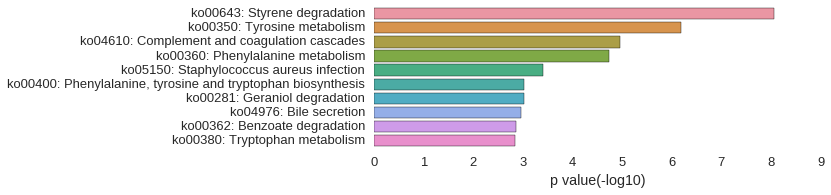
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04929
TCAAATGACAAGTATTCCTACTACATCACCTTCTGCACCTAACATGTCAGGAAATGTGACTAATGGGATG
TCGAACGCAACTTCTACTACAGCAAGTTCAATCACCAATTTGGGCAATGCAATGACAAGTGCAACCACCA
CAGCACCAACTGCACAACCTTTATTCAATCCTACAGCCCCACCTTCGTCCAATCCTACAGCACCACCTTC
GTCCAATCCTACAGCACCACCTTCATTCAATCCTACAGCCCCACCTTCGTCCAATCCTACAGCACCACCT
TCATTCAATTCTACAGCACCACCTCCATTCGTTTCTACAGCACCACCTCAAAATAACTCTGCTGCACCAC
CTTCATTCAATTCTACAGCACCACCTCCATTCGTTTCTACAGCACCACCTCAAAATAACTCTGCTGCACC
ACCTTCATTCAATTCTACAGCACCACCTCCATTCGTTTCTACAGCACCACCTCAAAATAACTCTGCTGCA
CCACCTTCATTCAATTCTACAGCACCACCTCCATTCAATTCTACAGCACCACCTCAAAATAACTCTGCTG
CACCACCTTCATTCGTTTCTACAGCACCACCTCAAAATAACTCTGCTGCACCACCTTCATTCGTTTCTAC
AGCACCACCTCAAAATAACTCTGCTGCACCACCTTCATTCAATTCTACAGCACCACCTCCATTCAATTCT
ATAGCACCACCTTCATTTAACTTTACAGCACCAGCTAGATTCAACTCCACAGCGCTACCTCCATTCAACT
CCACAGCACCACCTCCATTCAATTCTACAGCACCACCTTCATTTAACTCTACAGCACCAGCTGGATTCAA
CTCCACAGCGCCACCTCCATTCAATTCCACAGCACCACCTCCATTTGACTCTACCACAGGCTCTCCCAAA
GGTAATTCACTGTTAATTGTGTGTGTTAATATGTAGGGTCAGGTTTTAGCTTGAGTGTTAACAATCACTG
TGCATTTTACAGATGTTCTTACAATTCTGACTAATCTGGAGGTGGAGGTTTTGGCCGAAGAGTCGTCCTC
TGAAGAAGAACGTCAGGCTGCACTACGTCAGG
TCAAATGACAAGTATTCCTACTACATCACCTTCTGCACCTAACATGTCAGGAAATGTGACTAATGGGATG
TCGAACGCAACTTCTACTACAGCAAGTTCAATCACCAATTTGGGCAATGCAATGACAAGTGCAACCACCA
CAGCACCAACTGCACAACCTTTATTCAATCCTACAGCCCCACCTTCGTCCAATCCTACAGCACCACCTTC
GTCCAATCCTACAGCACCACCTTCATTCAATCCTACAGCCCCACCTTCGTCCAATCCTACAGCACCACCT
TCATTCAATTCTACAGCACCACCTCCATTCGTTTCTACAGCACCACCTCAAAATAACTCTGCTGCACCAC
CTTCATTCAATTCTACAGCACCACCTCCATTCGTTTCTACAGCACCACCTCAAAATAACTCTGCTGCACC
ACCTTCATTCAATTCTACAGCACCACCTCCATTCGTTTCTACAGCACCACCTCAAAATAACTCTGCTGCA
CCACCTTCATTCAATTCTACAGCACCACCTCCATTCAATTCTACAGCACCACCTCAAAATAACTCTGCTG
CACCACCTTCATTCGTTTCTACAGCACCACCTCAAAATAACTCTGCTGCACCACCTTCATTCGTTTCTAC
AGCACCACCTCAAAATAACTCTGCTGCACCACCTTCATTCAATTCTACAGCACCACCTCCATTCAATTCT
ATAGCACCACCTTCATTTAACTTTACAGCACCAGCTAGATTCAACTCCACAGCGCTACCTCCATTCAACT
CCACAGCACCACCTCCATTCAATTCTACAGCACCACCTTCATTTAACTCTACAGCACCAGCTGGATTCAA
CTCCACAGCGCCACCTCCATTCAATTCCACAGCACCACCTCCATTTGACTCTACCACAGGCTCTCCCAAA
GGTAATTCACTGTTAATTGTGTGTGTTAATATGTAGGGTCAGGTTTTAGCTTGAGTGTTAACAATCACTG
TGCATTTTACAGATGTTCTTACAATTCTGACTAATCTGGAGGTGGAGGTTTTGGCCGAAGAGTCGTCCTC
TGAAGAAGAACGTCAGGCTGCACTACGTCAGG