ZFLNCG04996
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1648856 | brain | normal | 147.66 |
| ERR023144 | brain | normal | 86.63 |
| SRR594771 | endothelium | normal | 77.97 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 68.84 |
| SRR372802 | 5 dpf | normal | 65.23 |
| SRR1648854 | brain | normal | 55.11 |
| SRR535978 | larvae | normal | 54.27 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 53.22 |
| SRR1562533 | testis | normal | 50.23 |
| SRR1569491 | embryo | normal | 49.23 |
Express in tissues
Correlated coding gene
Download
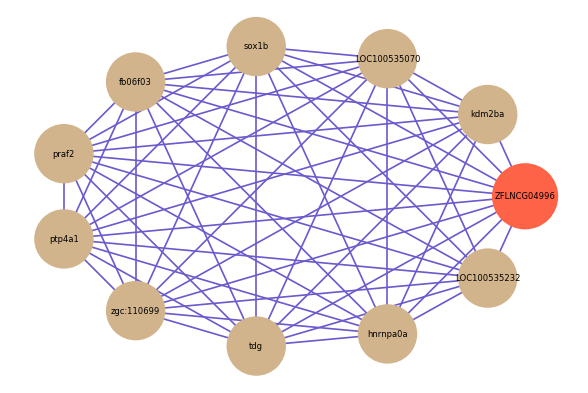
| gene | correlation coefficent |
|---|---|
| kdm2ba | 0.70 |
| LOC100535070 | 0.67 |
| LOC100535232 | 0.66 |
| sox1b | 0.64 |
| fb06f03 | 0.64 |
| praf2 | 0.63 |
| ptp4a1 | 0.63 |
| zgc:110699 | 0.63 |
| tdg | 0.62 |
| hnrnpa0a | 0.62 |
Gene Ontology
Download
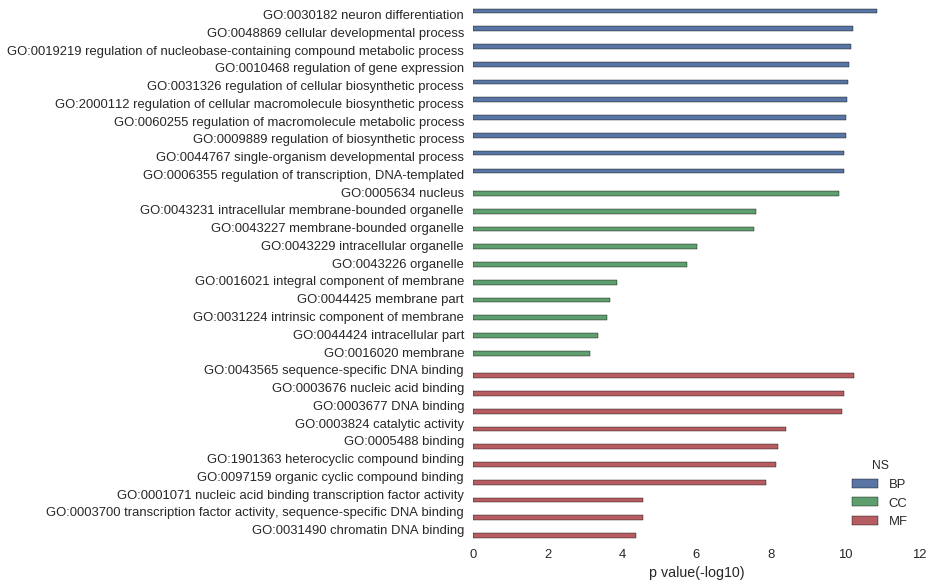
| GO | P value |
|---|---|
| GO:0030182 | 1.43e-11 |
| GO:0048869 | 6.06e-11 |
| GO:0019219 | 6.95e-11 |
| GO:0010468 | 7.98e-11 |
| GO:0031326 | 8.38e-11 |
| GO:2000112 | 8.76e-11 |
| GO:0060255 | 9.24e-11 |
| GO:0009889 | 9.47e-11 |
| GO:0044767 | 1.09e-10 |
| GO:0006355 | 1.10e-10 |
| GO:0005634 | 1.46e-10 |
| GO:0043231 | 2.45e-08 |
| GO:0043227 | 2.80e-08 |
| GO:0043229 | 9.57e-07 |
| GO:0043226 | 1.74e-06 |
| GO:0016021 | 1.39e-04 |
| GO:0044425 | 2.09e-04 |
| GO:0031224 | 2.54e-04 |
| GO:0044424 | 4.53e-04 |
| GO:0016020 | 7.46e-04 |
| GO:0043565 | 5.78e-11 |
| GO:0003676 | 1.09e-10 |
| GO:0003677 | 1.21e-10 |
| GO:0003824 | 3.91e-09 |
| GO:0005488 | 6.24e-09 |
| GO:1901363 | 7.41e-09 |
| GO:0097159 | 1.31e-08 |
| GO:0001071 | 2.64e-05 |
| GO:0003700 | 2.64e-05 |
| GO:0031490 | 4.10e-05 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04996
GGGTTAAAAGAGAGCTGAAATCCAGTCAGTAAAAGACTCTTTTTGTATTTGTATTGTGTATAGTTTGCTA
ACAATTGTGTATATGTGCCTGTACATATTGTGAAGGGGACGTGCTCCTTTTTTTTTTCTCCTCCAACTTG
GACAGAGTTACTGTCTCCATTCAGGAGAAATTGCGAGCCAAAATGTTGACATGAGTGAAATGGATCATTT
GTATCTGGAAACAAATAAAGCTATAATTTTAAC
GGGTTAAAAGAGAGCTGAAATCCAGTCAGTAAAAGACTCTTTTTGTATTTGTATTGTGTATAGTTTGCTA
ACAATTGTGTATATGTGCCTGTACATATTGTGAAGGGGACGTGCTCCTTTTTTTTTTCTCCTCCAACTTG
GACAGAGTTACTGTCTCCATTCAGGAGAAATTGCGAGCCAAAATGTTGACATGAGTGAAATGGATCATTT
GTATCTGGAAACAAATAAAGCTATAATTTTAAC