ZFLNCG05139
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 57.44 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 36.50 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 35.17 |
| ERR023144 | brain | normal | 33.26 |
| SRR514030 | retina | id2a morpholino | 15.20 |
| SRR514028 | retina | control morpholino | 14.87 |
| SRR1565809 | brain | normal | 14.39 |
| SRR1565805 | brain | normal | 13.18 |
| SRR1565811 | brain | normal | 11.76 |
| SRR1565807 | brain | normal | 11.09 |
Express in tissues
Correlated coding gene
Download
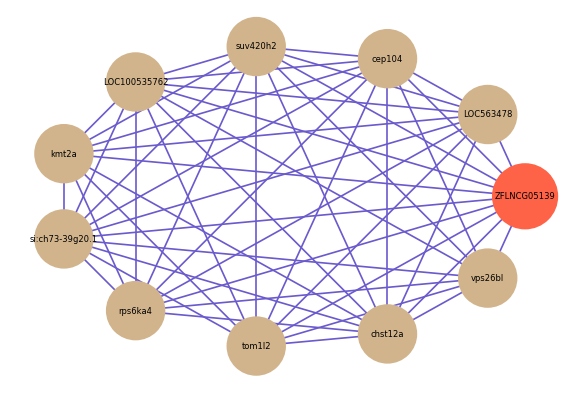
| gene | correlation coefficent |
|---|---|
| LOC563478 | 0.67 |
| cep104 | 0.63 |
| suv420h2 | 0.62 |
| vps26bl | 0.60 |
| LOC100535762 | 0.59 |
| kmt2a | 0.58 |
| si:ch73-39g20.1 | 0.57 |
| rps6ka4 | 0.57 |
| tom1l2 | 0.56 |
| chst12a | 0.56 |
Gene Ontology
Download
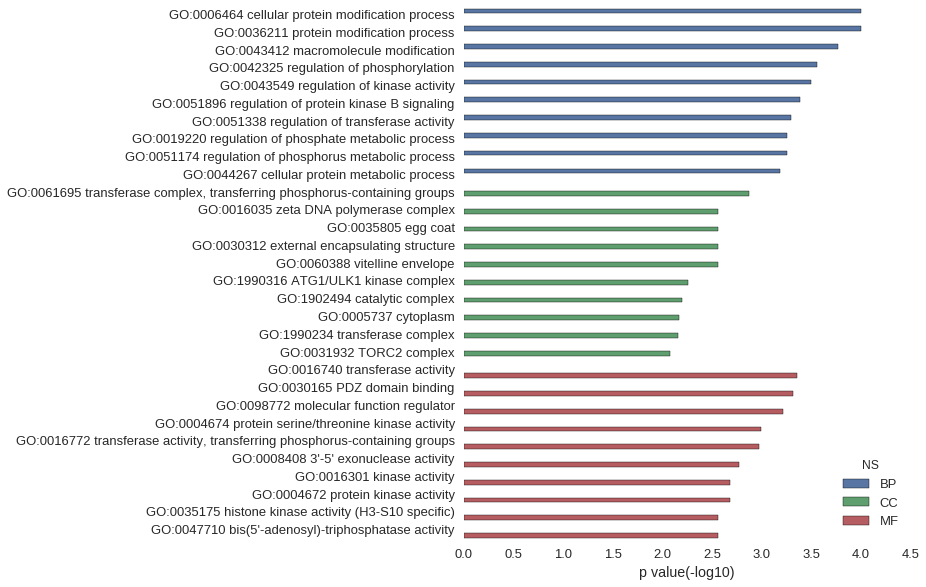
| GO | P value |
|---|---|
| GO:0006464 | 9.85e-05 |
| GO:0036211 | 9.85e-05 |
| GO:0043412 | 1.70e-04 |
| GO:0042325 | 2.75e-04 |
| GO:0043549 | 3.18e-04 |
| GO:0051896 | 4.05e-04 |
| GO:0051338 | 4.99e-04 |
| GO:0019220 | 5.55e-04 |
| GO:0051174 | 5.55e-04 |
| GO:0044267 | 6.53e-04 |
| GO:0061695 | 1.35e-03 |
| GO:0016035 | 2.77e-03 |
| GO:0035805 | 2.77e-03 |
| GO:0030312 | 2.77e-03 |
| GO:0060388 | 2.77e-03 |
| GO:1990316 | 5.54e-03 |
| GO:1902494 | 6.31e-03 |
| GO:0005737 | 6.72e-03 |
| GO:1990234 | 6.94e-03 |
| GO:0031932 | 8.29e-03 |
| GO:0016740 | 4.36e-04 |
| GO:0030165 | 4.85e-04 |
| GO:0098772 | 6.02e-04 |
| GO:0004674 | 1.01e-03 |
| GO:0016772 | 1.05e-03 |
| GO:0008408 | 1.67e-03 |
| GO:0016301 | 2.07e-03 |
| GO:0004672 | 2.08e-03 |
| GO:0035175 | 2.77e-03 |
| GO:0047710 | 2.77e-03 |
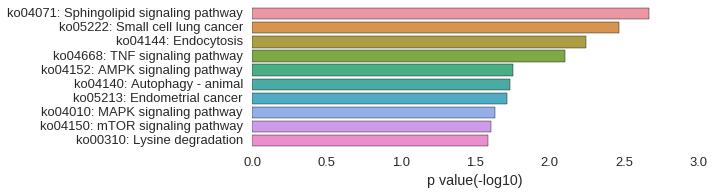
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05139
AAAAGACAGAAATCTCTCAGATTTCATTAAAAATATCTTAACTTGAGTTCCAAATATGAACATTTCCTTT
GAGTTTGGATTGACAAAAGGCTGAAGTAAATGACAGTTTTCATTTAGGGGTAAACAAACTTAGTTAAGGC
CAATTTACACTGCACTGACAGACAGAAACCAACACAGACAATACCCCAAATTACACTGCATCACTTCTCA
AACATTCCACGATCCAACAAGTGCCAACTAACACCTTCAAAAGCAACACGCTCCCCTTTTCCACCAAAGC
AGTTTAAGTGCTTCTTTAGAGCCAGAGCCTAGTTAAAGTTAGTTTTTGTTTCTTTACATTCAAAGCACCA
GTTCTAAACCAGAAATAATGGTTAATTAGCAGCATCAAAACATTACAGAGCTAATAGCAATAACCGTTTA
CATCAGGAACAGAAGCCGAAGTTACCCTACCAGTGTTTACCAACCACACAAACACAATCTTTAGAA
AAAAGACAGAAATCTCTCAGATTTCATTAAAAATATCTTAACTTGAGTTCCAAATATGAACATTTCCTTT
GAGTTTGGATTGACAAAAGGCTGAAGTAAATGACAGTTTTCATTTAGGGGTAAACAAACTTAGTTAAGGC
CAATTTACACTGCACTGACAGACAGAAACCAACACAGACAATACCCCAAATTACACTGCATCACTTCTCA
AACATTCCACGATCCAACAAGTGCCAACTAACACCTTCAAAAGCAACACGCTCCCCTTTTCCACCAAAGC
AGTTTAAGTGCTTCTTTAGAGCCAGAGCCTAGTTAAAGTTAGTTTTTGTTTCTTTACATTCAAAGCACCA
GTTCTAAACCAGAAATAATGGTTAATTAGCAGCATCAAAACATTACAGAGCTAATAGCAATAACCGTTTA
CATCAGGAACAGAAGCCGAAGTTACCCTACCAGTGTTTACCAACCACACAAACACAATCTTTAGAA