ZFLNCG05241
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 92.87 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 84.25 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 59.65 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 56.94 |
| SRR1004785 | larvae | normal | 24.20 |
| SRR801556 | embryo | RPS19 and p53 morpholino | 23.31 |
| SRR1708345 | embryo | SIBAS | 22.69 |
| SRR038625 | embryo | traf6 morpholino | 21.64 |
| SRR776510 | embryo | Salmonella infected and miR-146 knockdown | 18.91 |
| SRR1004787 | larvae | impdh1a morpholino | 17.94 |
Express in tissues
Correlated coding gene
Download
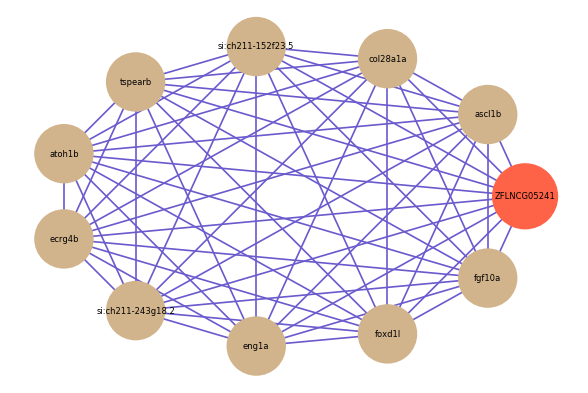
| gene | correlation coefficent |
|---|---|
| ascl1b | 0.80 |
| col28a1a | 0.79 |
| si:ch211-152f23.5 | 0.76 |
| tspearb | 0.76 |
| atoh1b | 0.75 |
| ecrg4b | 0.75 |
| si:ch211-243g18.2 | 0.75 |
| eng1a | 0.74 |
| foxd1l | 0.74 |
| fgf10a | 0.74 |
Gene Ontology
Download
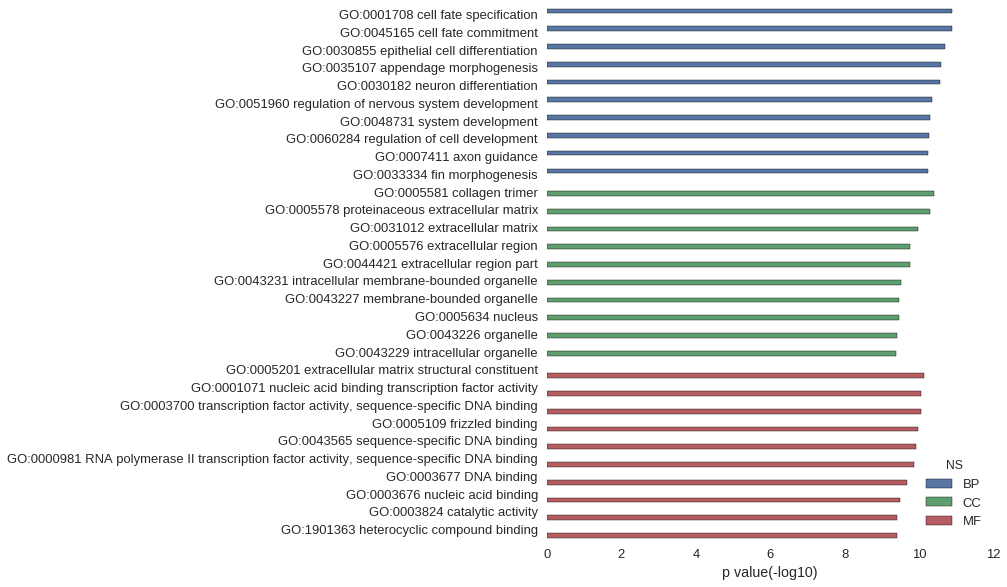
| GO | P value |
|---|---|
| GO:0001708 | 1.28e-11 |
| GO:0045165 | 1.28e-11 |
| GO:0030855 | 2.01e-11 |
| GO:0035107 | 2.62e-11 |
| GO:0030182 | 2.71e-11 |
| GO:0051960 | 4.60e-11 |
| GO:0048731 | 5.19e-11 |
| GO:0060284 | 5.26e-11 |
| GO:0007411 | 5.79e-11 |
| GO:0033334 | 5.80e-11 |
| GO:0005581 | 4.04e-11 |
| GO:0005578 | 5.23e-11 |
| GO:0031012 | 1.04e-10 |
| GO:0005576 | 1.72e-10 |
| GO:0044421 | 1.73e-10 |
| GO:0043231 | 3.10e-10 |
| GO:0043227 | 3.45e-10 |
| GO:0005634 | 3.45e-10 |
| GO:0043226 | 3.94e-10 |
| GO:0043229 | 4.08e-10 |
| GO:0005201 | 7.41e-11 |
| GO:0001071 | 8.66e-11 |
| GO:0003700 | 8.66e-11 |
| GO:0005109 | 1.07e-10 |
| GO:0043565 | 1.21e-10 |
| GO:0000981 | 1.38e-10 |
| GO:0003677 | 2.06e-10 |
| GO:0003676 | 3.29e-10 |
| GO:0003824 | 3.83e-10 |
| GO:1901363 | 3.94e-10 |
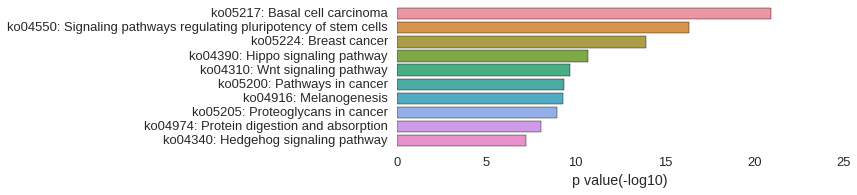
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05241
CCCTTCACCACAGGTGTAAGAAGAGAAATCTTCCCAGGCAAAATATCCGTCCCTGTATCCATAGCCACAA
ACTTTCCTCCCAAAAGCACAAACCAACAGAATGGATGGAATGAGCACGGATACAAGAGAGGTGGTTGAAC
TCGACGTCCAGCATTCGAGCTTGGGGCGGGGGGAGCAGAGCGAGTACCCACCAGCCTTGGCACTCATGGC
CAGCAGTGACCCACGCGCCTGGCTGGCTCCCGTGCAGGCTGGCACCTGCGCGGCACACGCCGAATACCTG
CTGCACTCGCCCGGCTCGAGCGCGGAAGGCGTGTCCTCTGCCTCCAACTTCAGGAAGAGCAGCAAGAGTC
CTGTCAAAGTACGCGAGCTCTGCCGGCTTAAAGGAGCTGTGGGGGCAGATGAGGGCAGACAGCGGGCCCC
ATCCAGCAAATCCACCAACGTCGTGCAGAAACAGAGGCGAATGGCTGCCAATGCCCGGGAGAGGCGAAGA
ATGCACGGATTGAACCACGCGTTCGACGAGCTGCGCAGTGTCATCCCAGCCTTTGACAACGACAAGAAAC
TCTCCAAGTACGAAACCCTGCAGATGGCCCAGATCTACATCAACGCCCTGTCCGACTTACTACAGGGCCC
CGGTGCTAAAGCCGACCCGCCAAACTGCGACCTGCTGCATGCCAACGTGTTAGAAACGGACCGATCTCCC
AGAGGATCACCGGGCGTCTGTCGGAGAGGCACGGGCGTGGGTTACCCGTACCAGTACGAGGACGGAACAT
TCAACTCTTTCATGGAGCAAGACCTCCAGTCGCCCTCTGGAACGAGCAAGTCTGGTTCGGAGGCCAGTAA
AGACTCGCCTCGGTCGAACCGGAGTGATGGAGAGTTCTCGCCTCACTC
CCCTTCACCACAGGTGTAAGAAGAGAAATCTTCCCAGGCAAAATATCCGTCCCTGTATCCATAGCCACAA
ACTTTCCTCCCAAAAGCACAAACCAACAGAATGGATGGAATGAGCACGGATACAAGAGAGGTGGTTGAAC
TCGACGTCCAGCATTCGAGCTTGGGGCGGGGGGAGCAGAGCGAGTACCCACCAGCCTTGGCACTCATGGC
CAGCAGTGACCCACGCGCCTGGCTGGCTCCCGTGCAGGCTGGCACCTGCGCGGCACACGCCGAATACCTG
CTGCACTCGCCCGGCTCGAGCGCGGAAGGCGTGTCCTCTGCCTCCAACTTCAGGAAGAGCAGCAAGAGTC
CTGTCAAAGTACGCGAGCTCTGCCGGCTTAAAGGAGCTGTGGGGGCAGATGAGGGCAGACAGCGGGCCCC
ATCCAGCAAATCCACCAACGTCGTGCAGAAACAGAGGCGAATGGCTGCCAATGCCCGGGAGAGGCGAAGA
ATGCACGGATTGAACCACGCGTTCGACGAGCTGCGCAGTGTCATCCCAGCCTTTGACAACGACAAGAAAC
TCTCCAAGTACGAAACCCTGCAGATGGCCCAGATCTACATCAACGCCCTGTCCGACTTACTACAGGGCCC
CGGTGCTAAAGCCGACCCGCCAAACTGCGACCTGCTGCATGCCAACGTGTTAGAAACGGACCGATCTCCC
AGAGGATCACCGGGCGTCTGTCGGAGAGGCACGGGCGTGGGTTACCCGTACCAGTACGAGGACGGAACAT
TCAACTCTTTCATGGAGCAAGACCTCCAGTCGCCCTCTGGAACGAGCAAGTCTGGTTCGGAGGCCAGTAA
AGACTCGCCTCGGTCGAACCGGAGTGATGGAGAGTTCTCGCCTCACTC