ZFLNCG05312
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562529 | intestine and pancreas | normal | 22.03 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 21.62 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 18.94 |
| SRR726542 | 5 dpf | infection with control | 14.23 |
| ERR023146 | head kidney | normal | 12.17 |
| SRR726540 | 5 dpf | normal | 10.34 |
| SRR633516 | larvae | normal | 9.82 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 8.76 |
| SRR1035239 | liver | transgenic mCherry control | 8.16 |
| SRR633550 | larvae | exposed to hypoxia stress | 7.19 |
Express in tissues
Correlated coding gene
Download
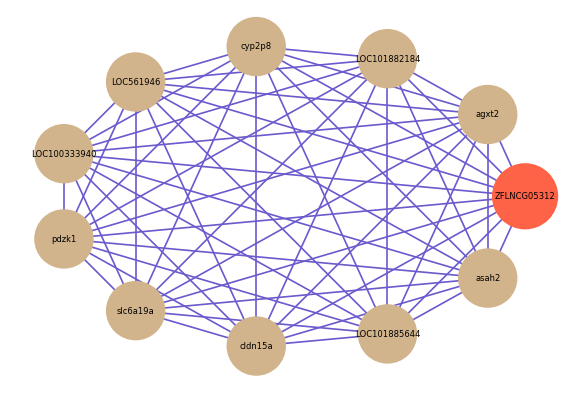
| gene | correlation coefficent |
|---|---|
| agxt2 | 0.68 |
| LOC101882184 | 0.67 |
| cyp2p8 | 0.66 |
| LOC561946 | 0.66 |
| LOC100333940 | 0.66 |
| pdzk1 | 0.66 |
| slc6a19a | 0.66 |
| cldn15a | 0.65 |
| LOC101885644 | 0.65 |
| asah2 | 0.65 |
Gene Ontology
Download
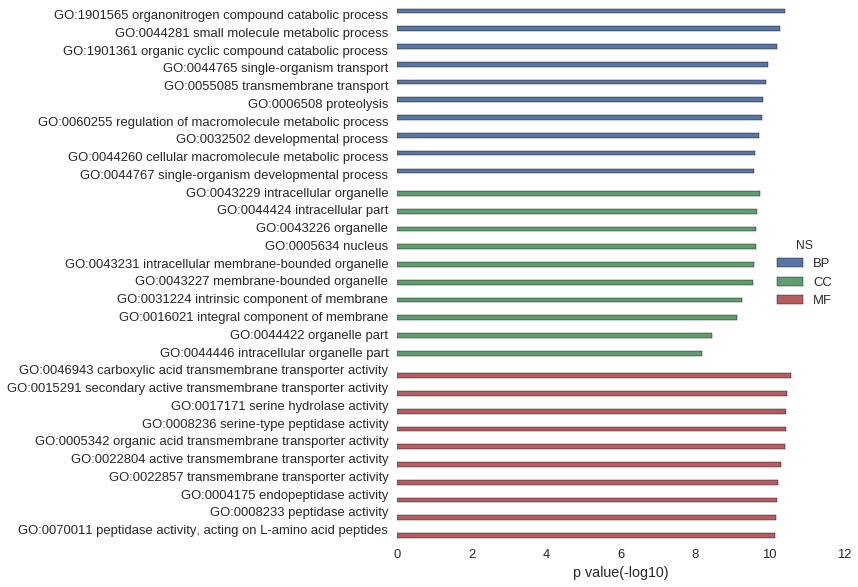
| GO | P value |
|---|---|
| GO:1901565 | 3.92e-11 |
| GO:0044281 | 5.24e-11 |
| GO:1901361 | 6.29e-11 |
| GO:0044765 | 1.08e-10 |
| GO:0055085 | 1.22e-10 |
| GO:0006508 | 1.45e-10 |
| GO:0060255 | 1.63e-10 |
| GO:0032502 | 1.94e-10 |
| GO:0044260 | 2.38e-10 |
| GO:0044767 | 2.60e-10 |
| GO:0043229 | 1.79e-10 |
| GO:0044424 | 2.20e-10 |
| GO:0043226 | 2.32e-10 |
| GO:0005634 | 2.34e-10 |
| GO:0043231 | 2.64e-10 |
| GO:0043227 | 2.79e-10 |
| GO:0031224 | 5.50e-10 |
| GO:0016021 | 7.41e-10 |
| GO:0044422 | 3.42e-09 |
| GO:0044446 | 6.57e-09 |
| GO:0046943 | 2.61e-11 |
| GO:0015291 | 3.38e-11 |
| GO:0017171 | 3.54e-11 |
| GO:0008236 | 3.54e-11 |
| GO:0005342 | 3.70e-11 |
| GO:0022804 | 4.98e-11 |
| GO:0022857 | 5.72e-11 |
| GO:0004175 | 6.28e-11 |
| GO:0008233 | 6.51e-11 |
| GO:0070011 | 7.06e-11 |
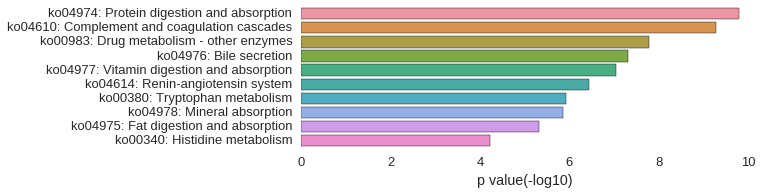
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05312
CAGGCTTGGTTTACAGGTAACACAATCATTGCTCTGATGTCCATTACATGATGCACAGTTCAAATGACAC
TGAACACACTCTTGTTTTTCCTTGTCTTCAAAGTAACCATCCGGACACACGGACACACAAGTCTGATCTG
ATAAAAATAACCAAATATGCACAATGTCAAATCTATAATTTATATTTAAGGTTTAGTTTTTACAAACTTT
TTAATTTTGTTCATGTTAATTTTAGTTCATGTTAATAATTTTAATTTAAGACATTTATATTTGGTGAGGT
CTCACTGAGCAAGATGTGATTGGGTTTGCAGCTGAGGCATTGATCCTTATCTGTGCCAGAACAGTGGAGA
CAGCTCTTGTGACATGGCTGGCCCTGACCACTTTCATCCATGAATGTAGAGCAGTCACTGACAAATTCAC
AGTGACCATTATCATCTATCTTCATGTTCTCTCTGCAGGTGAGGCATGAGGACAGGTAAGGACCTGAGCA
AGTCAAGCATGACTTGTCACAGTCTA
CAGGCTTGGTTTACAGGTAACACAATCATTGCTCTGATGTCCATTACATGATGCACAGTTCAAATGACAC
TGAACACACTCTTGTTTTTCCTTGTCTTCAAAGTAACCATCCGGACACACGGACACACAAGTCTGATCTG
ATAAAAATAACCAAATATGCACAATGTCAAATCTATAATTTATATTTAAGGTTTAGTTTTTACAAACTTT
TTAATTTTGTTCATGTTAATTTTAGTTCATGTTAATAATTTTAATTTAAGACATTTATATTTGGTGAGGT
CTCACTGAGCAAGATGTGATTGGGTTTGCAGCTGAGGCATTGATCCTTATCTGTGCCAGAACAGTGGAGA
CAGCTCTTGTGACATGGCTGGCCCTGACCACTTTCATCCATGAATGTAGAGCAGTCACTGACAAATTCAC
AGTGACCATTATCATCTATCTTCATGTTCTCTCTGCAGGTGAGGCATGAGGACAGGTAAGGACCTGAGCA
AGTCAAGCATGACTTGTCACAGTCTA