ZFLNCG05365
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR527832 | 16-36 hpf | normal | 37.57 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 17.48 |
| SRR658547 | 6 somite | Gata5/6 morphant | 16.25 |
| SRR658543 | 6 somite | Gata5 morphan | 15.28 |
| SRR038627 | embryo | control morpholino | 13.71 |
| SRR516124 | skin | male and 3.5 year | 12.37 |
| SRR658545 | 6 somite | Gata6 morphant | 11.61 |
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 11.30 |
| SRR1021215 | sphere stage | Mzeomesa | 10.92 |
| SRR726542 | 5 dpf | infection with control | 10.41 |
Express in tissues
Gene Ontology
Download
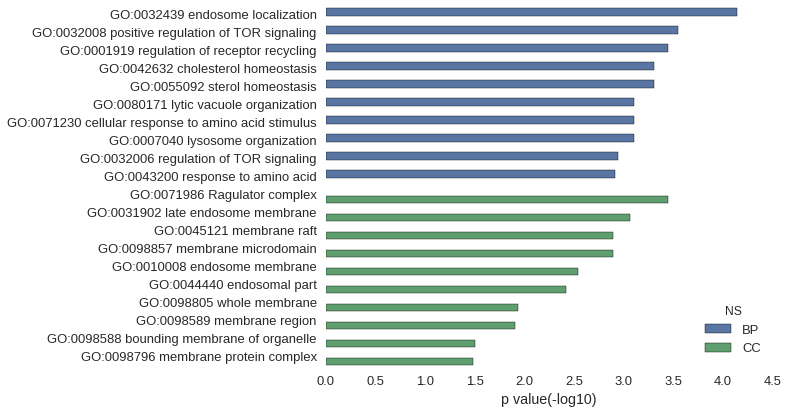
| GO | P value |
|---|---|
| GO:0032439 | 7.11e-05 |
| GO:0032008 | 2.84e-04 |
| GO:0001919 | 3.55e-04 |
| GO:0042632 | 4.97e-04 |
| GO:0055092 | 4.97e-04 |
| GO:0080171 | 7.82e-04 |
| GO:0071230 | 7.82e-04 |
| GO:0007040 | 7.82e-04 |
| GO:0032006 | 1.14e-03 |
| GO:0043200 | 1.21e-03 |
| GO:0071986 | 3.55e-04 |
| GO:0031902 | 8.53e-04 |
| GO:0045121 | 1.28e-03 |
| GO:0098857 | 1.28e-03 |
| GO:0010008 | 2.91e-03 |
| GO:0044440 | 3.77e-03 |
| GO:0098805 | 1.17e-02 |
| GO:0098589 | 1.24e-02 |
| GO:0098588 | 3.15e-02 |
| GO:0098796 | 3.27e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05365
TTTCTTTCGCAGAGGTCAATTGTAAACCACTGTCATTTAACGATAGTTTTTGTTCTCTGCTGAAAGGTGG
AGATGTGGTGAAGGTTGCTCAGCTGGTGGGTCAGAGTCGGATCGCTCAGCCGGAGACTCCAGTGACCCTG
CAGTTTCAGGGAAACAAGTTCACCTTGTCCCAGAGTCAGCTGCGTCAGCTCACCACCGGCCAACCCCTGC
AGCTTCAAGGTACACTCGGTAATGTCCAGCTCATATTCATCTTCTCTCTGTCATTTCACCTCCATATATA
GTCAGAATTGTGCGTTTATTTATTTATAATGCTCATGTCTATAAATGTCTGCTTTAGGTAACATCCTGCA
GATTGTGTCAGCTCCTGGACAGCCTCTTCTCAGGCCTCAGGGGCCACCTATGG
TTTCTTTCGCAGAGGTCAATTGTAAACCACTGTCATTTAACGATAGTTTTTGTTCTCTGCTGAAAGGTGG
AGATGTGGTGAAGGTTGCTCAGCTGGTGGGTCAGAGTCGGATCGCTCAGCCGGAGACTCCAGTGACCCTG
CAGTTTCAGGGAAACAAGTTCACCTTGTCCCAGAGTCAGCTGCGTCAGCTCACCACCGGCCAACCCCTGC
AGCTTCAAGGTACACTCGGTAATGTCCAGCTCATATTCATCTTCTCTCTGTCATTTCACCTCCATATATA
GTCAGAATTGTGCGTTTATTTATTTATAATGCTCATGTCTATAAATGTCTGCTTTAGGTAACATCCTGCA
GATTGTGTCAGCTCCTGGACAGCCTCTTCTCAGGCCTCAGGGGCCACCTATGG