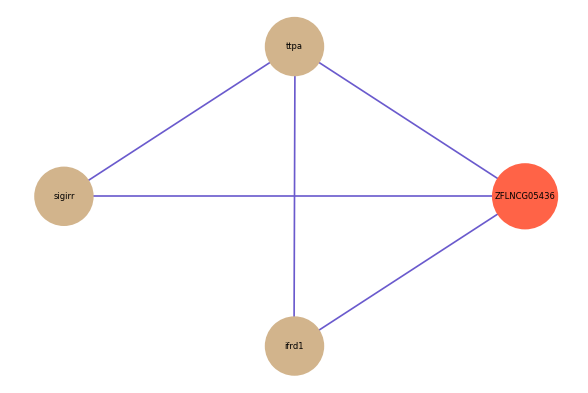
ZFLNCG05436
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035239 | liver | transgenic mCherry control | 19.70 |
| SRR891504 | liver | normal | 10.88 |
| ERR023143 | swim bladder | normal | 10.63 |
| SRR1299129 | caudal fin | Seven days time after treatment | 7.30 |
| SRR1299128 | caudal fin | Three days time after treatment | 6.78 |
| SRR1562530 | liver | normal | 5.76 |
| SRR1647682 | spleen | normal | 5.13 |
| SRR1299127 | caudal fin | Two days time after treatment | 4.78 |
| SRR1167762 | embryo | ptpn6 morpholino | 4.66 |
| SRR065197 | 3 dpf | U1C knockout | 4.62 |
Express in tissues
Gene Ontology
Download
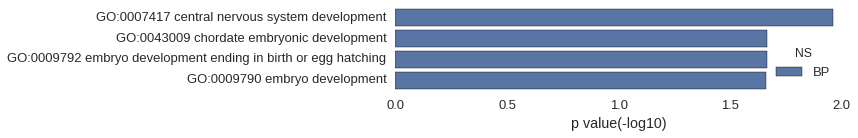
| GO | P value |
|---|---|
| GO:0007417 | 1.10e-02 |
| GO:0043009 | 2.16e-02 |
| GO:0009792 | 2.16e-02 |
| GO:0009790 | 2.18e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05436
TCGATCAACAGATTTTTTAACAGTGTATATACAGTAATTCTGAGGCAAAACAAACATCATCATCCTACAT
TTATTTTTTTTTTTCGGCGTGCTAGTCCTCTAACCCACCTCAAACCTAACTTTATTTAGCTCTCCCCATC
AATTCTGCTTAATTTTTAATGCTGGGAAGGGTAGATAGGCATTAATAATGCACTGTGGTGACCATTCATA
ATTGATGAACCGTTATAGAGCAAATCCAACACTGATACCCGGTTTATTTAGAAGTCATCAGAACGATCAA
TCTTAAATGTGAGCGCAAGACCTTGAGAAACCATCAACAAATAATAAAACGAACTCCTTTTCCCTTCGGC
GTTCGGCTAAAATTAGAGAAAAAACATCACTGAAAGCATTTAAAGGTGAGTTTAATTAGCTTTCGTACAT
ATCGTGCTAACTACTGGACTTGAATTGATATATTTGGGCTTTTATTCAATTAAACGGTCAGTCCATAATT
CCTGAGACTACAAGACAAATGTGACATGAAG
TCGATCAACAGATTTTTTAACAGTGTATATACAGTAATTCTGAGGCAAAACAAACATCATCATCCTACAT
TTATTTTTTTTTTTCGGCGTGCTAGTCCTCTAACCCACCTCAAACCTAACTTTATTTAGCTCTCCCCATC
AATTCTGCTTAATTTTTAATGCTGGGAAGGGTAGATAGGCATTAATAATGCACTGTGGTGACCATTCATA
ATTGATGAACCGTTATAGAGCAAATCCAACACTGATACCCGGTTTATTTAGAAGTCATCAGAACGATCAA
TCTTAAATGTGAGCGCAAGACCTTGAGAAACCATCAACAAATAATAAAACGAACTCCTTTTCCCTTCGGC
GTTCGGCTAAAATTAGAGAAAAAACATCACTGAAAGCATTTAAAGGTGAGTTTAATTAGCTTTCGTACAT
ATCGTGCTAACTACTGGACTTGAATTGATATATTTGGGCTTTTATTCAATTAAACGGTCAGTCCATAATT
CCTGAGACTACAAGACAAATGTGACATGAAG