ZFLNCG05461
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR658537 | bud | Gata6 morphant | 127.23 |
| SRR658539 | bud | Gata5/6 morphant | 86.60 |
| SRR658533 | bud | normal | 63.45 |
| SRR658535 | bud | Gata5 morphant | 60.57 |
| SRR658541 | 6 somite | normal | 57.32 |
| SRR658547 | 6 somite | Gata5/6 morphant | 46.83 |
| SRR749514 | 24 hpf | normal | 45.02 |
| SRR658543 | 6 somite | Gata5 morphan | 43.89 |
| SRR658545 | 6 somite | Gata6 morphant | 42.07 |
| SRR891504 | liver | normal | 41.61 |
Express in tissues
Correlated coding gene
Download
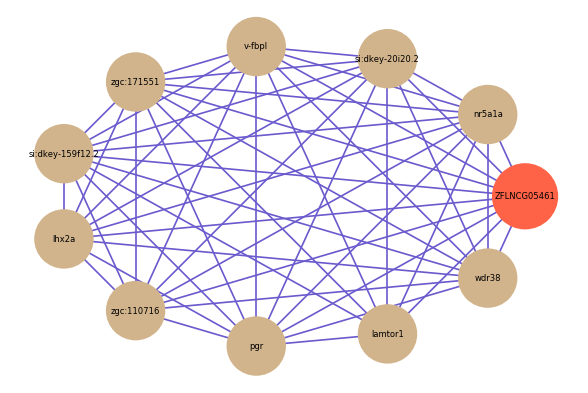
| gene | correlation coefficent |
|---|---|
| nr5a1a | 0.68 |
| si:dkey-20i20.2 | 0.65 |
| v-fbpl | 0.65 |
| zgc:171551 | 0.62 |
| si:dkey-159f12.2 | 0.61 |
| lhx2a | 0.60 |
| zgc:110716 | 0.60 |
| pgr | 0.59 |
| lamtor1 | 0.58 |
| wdr38 | 0.58 |
Gene Ontology
Download
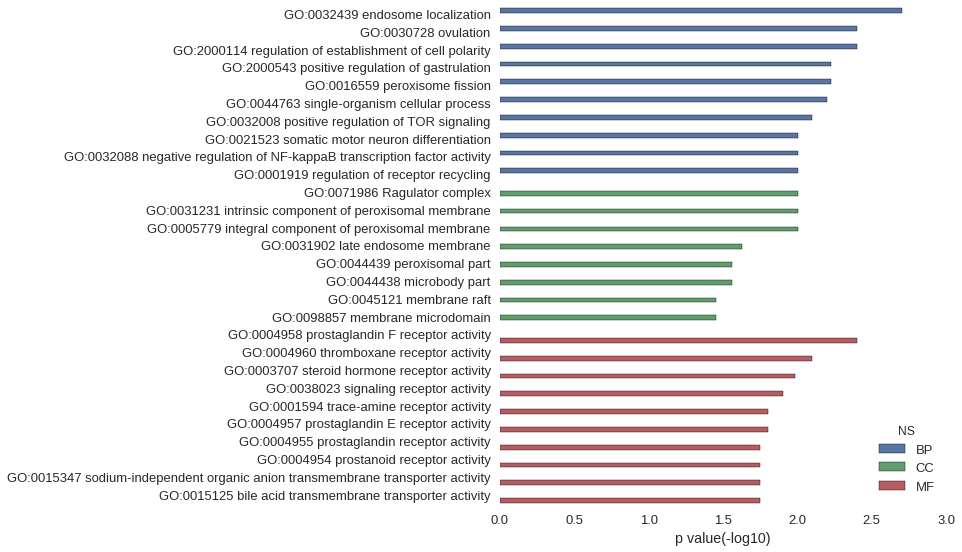
| GO | P value |
|---|---|
| GO:0032439 | 1.99e-03 |
| GO:0030728 | 3.98e-03 |
| GO:2000114 | 3.98e-03 |
| GO:2000543 | 5.96e-03 |
| GO:0016559 | 5.96e-03 |
| GO:0044763 | 6.35e-03 |
| GO:0032008 | 7.94e-03 |
| GO:0021523 | 9.91e-03 |
| GO:0032088 | 9.91e-03 |
| GO:0001919 | 9.91e-03 |
| GO:0071986 | 9.91e-03 |
| GO:0031231 | 9.91e-03 |
| GO:0005779 | 9.91e-03 |
| GO:0031902 | 2.36e-02 |
| GO:0044439 | 2.75e-02 |
| GO:0044438 | 2.75e-02 |
| GO:0045121 | 3.52e-02 |
| GO:0098857 | 3.52e-02 |
| GO:0004958 | 3.98e-03 |
| GO:0004960 | 7.94e-03 |
| GO:0003707 | 1.04e-02 |
| GO:0038023 | 1.25e-02 |
| GO:0001594 | 1.58e-02 |
| GO:0004957 | 1.58e-02 |
| GO:0004955 | 1.78e-02 |
| GO:0004954 | 1.78e-02 |
| GO:0015347 | 1.78e-02 |
| GO:0015125 | 1.78e-02 |
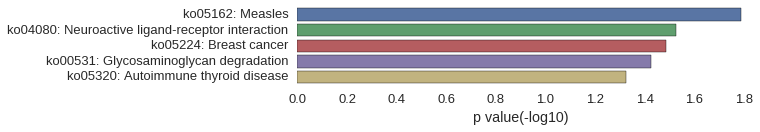
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05461
CGCCTGCAGAGCTTCTTCCCTTCTTCCTCATCCGCGGCTTGAATTTCTGTTTCGCCAAAGGATCAAATCC
CTGAAACAGACAAGACAAAGTCAACACTAATGTTCCTTTTATACTATGGTTATGGTAAACACTCACTCGA
GTACAATCACACTATATTTACATTCAGATGTGTGTGTCAGCTTAAATGACATGCAGTTCACACACACTAT
TATGTGATGTATATCGTACAAACTTTGAAGGACATTTTAACACTCGTGCAACAAGCAGATGACAGTTCAA
ACAATCACAGTCATCAGTTTGATTAAAAATCACCATTAATAGCGTCAGGCCGACTCACCATTACTTCAAC
CCGTTCCTTGTGTCTCTGCTCAAAGCTGGCCTGGTCCACTTTACTCAGCAGTCCGGGATCCAGACCGATC
AGCTCGGGCTGCACTTTCTCCAGCAGAGCCTTGACCTCCCACTCCTGCCTCTGTTTAGCGCTGCGGTACG
GGTTCGTATCGAGAGCGTCGAAATTCGGTTCTCCTGCAC
CGCCTGCAGAGCTTCTTCCCTTCTTCCTCATCCGCGGCTTGAATTTCTGTTTCGCCAAAGGATCAAATCC
CTGAAACAGACAAGACAAAGTCAACACTAATGTTCCTTTTATACTATGGTTATGGTAAACACTCACTCGA
GTACAATCACACTATATTTACATTCAGATGTGTGTGTCAGCTTAAATGACATGCAGTTCACACACACTAT
TATGTGATGTATATCGTACAAACTTTGAAGGACATTTTAACACTCGTGCAACAAGCAGATGACAGTTCAA
ACAATCACAGTCATCAGTTTGATTAAAAATCACCATTAATAGCGTCAGGCCGACTCACCATTACTTCAAC
CCGTTCCTTGTGTCTCTGCTCAAAGCTGGCCTGGTCCACTTTACTCAGCAGTCCGGGATCCAGACCGATC
AGCTCGGGCTGCACTTTCTCCAGCAGAGCCTTGACCTCCCACTCCTGCCTCTGTTTAGCGCTGCGGTACG
GGTTCGTATCGAGAGCGTCGAAATTCGGTTCTCCTGCAC