ZFLNCG05465
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR514028 | retina | control morpholino | 23.80 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 9.54 |
| SRR726542 | 5 dpf | infection with control | 9.22 |
| SRR592700 | pineal gland | normal | 7.33 |
| ERR023144 | brain | normal | 6.84 |
| SRR372800 | 2 dpf | normal | 5.66 |
| SRR038627 | embryo | control morpholino | 5.04 |
| ERR023143 | swim bladder | normal | 4.93 |
| SRR065196 | 3 dpf | normal | 4.56 |
| SRR519752 | 7 dpf | VD3 treatment | 4.24 |
Express in tissues
Correlated coding gene
Download
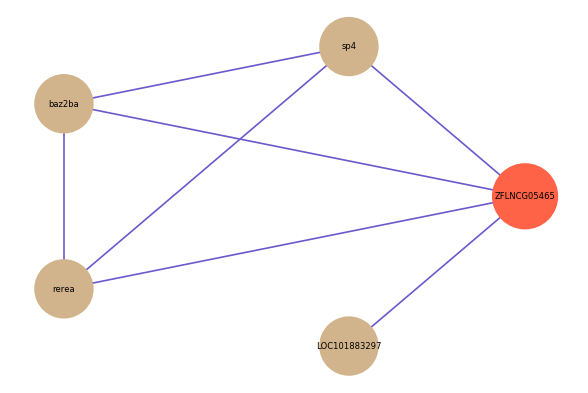
| gene | correlation coefficent |
|---|---|
| sp4 | 0.55 |
| LOC101883297 | 0.52 |
| baz2ba | 0.51 |
| rerea | 0.50 |
Gene Ontology
Download
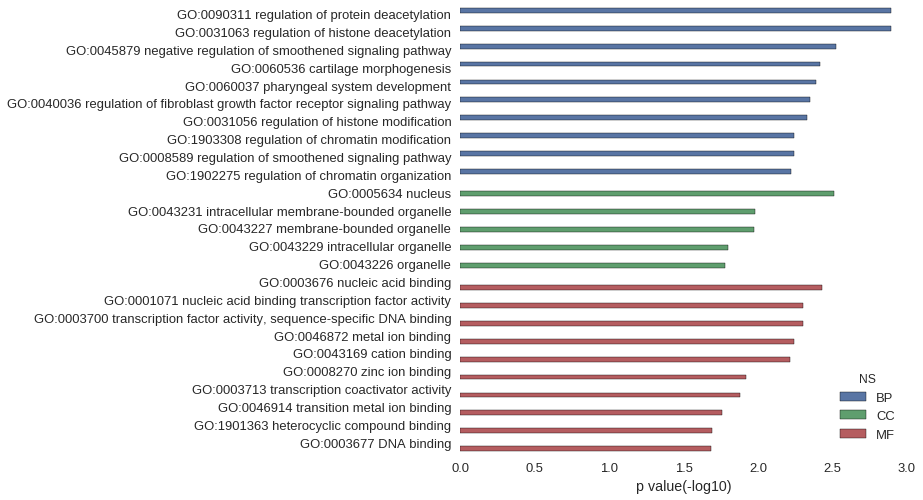
| GO | P value |
|---|---|
| GO:0090311 | 1.28e-03 |
| GO:0031063 | 1.28e-03 |
| GO:0045879 | 2.98e-03 |
| GO:0060536 | 3.83e-03 |
| GO:0060037 | 4.05e-03 |
| GO:0040036 | 4.47e-03 |
| GO:0031056 | 4.68e-03 |
| GO:1903308 | 5.75e-03 |
| GO:0008589 | 5.75e-03 |
| GO:1902275 | 5.96e-03 |
| GO:0005634 | 3.11e-03 |
| GO:0043231 | 1.05e-02 |
| GO:0043227 | 1.07e-02 |
| GO:0043229 | 1.59e-02 |
| GO:0043226 | 1.67e-02 |
| GO:0003676 | 3.69e-03 |
| GO:0001071 | 5.00e-03 |
| GO:0003700 | 5.00e-03 |
| GO:0046872 | 5.74e-03 |
| GO:0043169 | 6.10e-03 |
| GO:0008270 | 1.21e-02 |
| GO:0003713 | 1.32e-02 |
| GO:0046914 | 1.75e-02 |
| GO:1901363 | 2.03e-02 |
| GO:0003677 | 2.07e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05465
GCTTCAATGTGCGAGTGTGTGTGCGCGTTTTTGAATTCTGAATTCTTTTTGAATGCGTGTGTGTGAGAGT
GTGTGCTTGCATGCATGTTTGCACAAGTGTACACATGTGACTCTGCACGTCGTGTGTGTTTTGAGTGCTT
GTGTGAGTGTATGCATGTTCATGTGTATGCGCATGCGAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGT
GTGTGTGTGTGTGTGTGTGTGTGTGTGTATGCGTTAAGTGCTTGTGAGTCCATGTGTGTGTGCAGGTGAA
TCCCCAAACCTCTGTGAGCCTCAAAGCAGCTTCTGTTATCAAACTACACACACACACACACACACACACA
CACAGCGGTGGACAGTAGTCGGGACACACATGATCAGAACACACTAATGTCTTCATGGATCTCTGTACAG
TGTGAAGACATCGCTCTGTTAAGAAGCTGATTCAGGCCATCAAACATCTGGATTGTAAAG
GCTTCAATGTGCGAGTGTGTGTGCGCGTTTTTGAATTCTGAATTCTTTTTGAATGCGTGTGTGTGAGAGT
GTGTGCTTGCATGCATGTTTGCACAAGTGTACACATGTGACTCTGCACGTCGTGTGTGTTTTGAGTGCTT
GTGTGAGTGTATGCATGTTCATGTGTATGCGCATGCGAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGT
GTGTGTGTGTGTGTGTGTGTGTGTGTGTATGCGTTAAGTGCTTGTGAGTCCATGTGTGTGTGCAGGTGAA
TCCCCAAACCTCTGTGAGCCTCAAAGCAGCTTCTGTTATCAAACTACACACACACACACACACACACACA
CACAGCGGTGGACAGTAGTCGGGACACACATGATCAGAACACACTAATGTCTTCATGGATCTCTGTACAG
TGTGAAGACATCGCTCTGTTAAGAAGCTGATTCAGGCCATCAAACATCTGGATTGTAAAG

