ZFLNCG05483
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR941753 | posterior pectoral fin | normal | 39.81 |
| SRR941749 | anterior pectoral fin | normal | 38.73 |
| SRR527834 | head | normal | 35.67 |
| SRR1048059 | pineal gland | light | 24.19 |
| SRR1299124 | caudal fin | Zero day time after treatment | 22.63 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 21.68 |
| SRR1028004 | head | normal | 16.18 |
| SRR516126 | skin | male and 5 month | 14.02 |
| SRR516134 | skin | male and 5 month | 12.32 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 11.01 |
Express in tissues
Correlated coding gene
Download
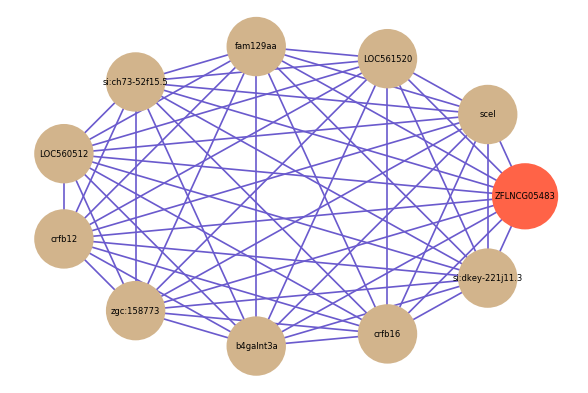
| gene | correlation coefficent |
|---|---|
| scel | 0.60 |
| LOC561520 | 0.59 |
| fam129aa | 0.59 |
| si:ch73-52f15.5 | 0.59 |
| LOC560512 | 0.58 |
| crfb12 | 0.58 |
| zgc:158773 | 0.58 |
| b4galnt3a | 0.57 |
| crfb16 | 0.57 |
| si:dkey-221j11.3 | 0.57 |
Gene Ontology
Download
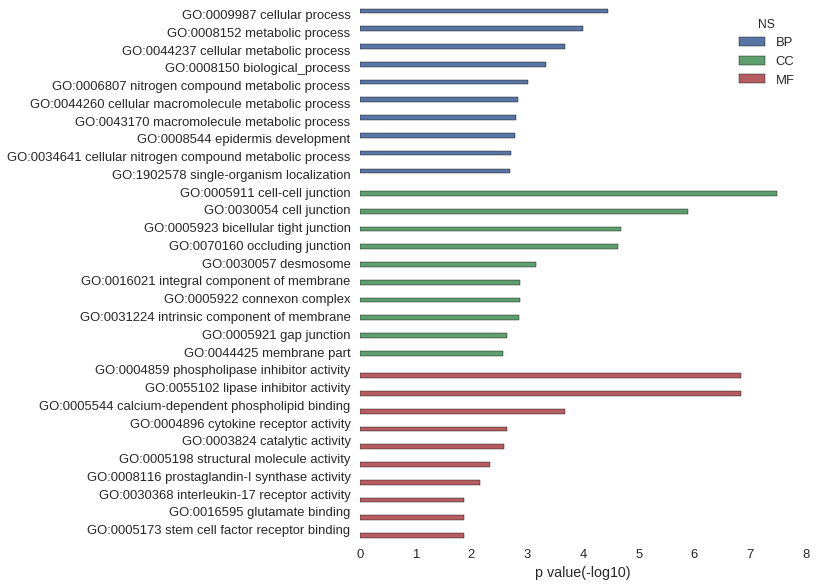
| GO | P value |
|---|---|
| GO:0009987 | 3.51e-05 |
| GO:0008152 | 9.96e-05 |
| GO:0044237 | 2.09e-04 |
| GO:0008150 | 4.53e-04 |
| GO:0006807 | 9.49e-04 |
| GO:0044260 | 1.47e-03 |
| GO:0043170 | 1.57e-03 |
| GO:0008544 | 1.64e-03 |
| GO:0034641 | 1.98e-03 |
| GO:1902578 | 2.02e-03 |
| GO:0005911 | 3.37e-08 |
| GO:0030054 | 1.30e-06 |
| GO:0005923 | 2.10e-05 |
| GO:0070160 | 2.32e-05 |
| GO:0030057 | 6.93e-04 |
| GO:0016021 | 1.33e-03 |
| GO:0005922 | 1.36e-03 |
| GO:0031224 | 1.39e-03 |
| GO:0005921 | 2.25e-03 |
| GO:0044425 | 2.66e-03 |
| GO:0004859 | 1.45e-07 |
| GO:0055102 | 1.45e-07 |
| GO:0005544 | 2.13e-04 |
| GO:0004896 | 2.25e-03 |
| GO:0003824 | 2.56e-03 |
| GO:0005198 | 4.70e-03 |
| GO:0008116 | 6.89e-03 |
| GO:0030368 | 1.37e-02 |
| GO:0016595 | 1.37e-02 |
| GO:0005173 | 1.37e-02 |
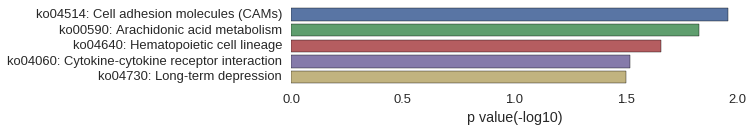
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05483
TCGACAATACCTGTCCTCTCAGAGAAACAGGGGCACAGACAGTAATGGAGTAAACCTCTTACAGGAGGCT
CGTCTTCATACAGCACACCTAATCATGATGTATTTACAGTAAACTAGCACATATTACAGCCTACACTGCT
GGAACACACGTCAACAGCTGGACCAACCGGTTTATACACTGAGAGAAAGAGAAAGTGTGAAAATGAGAAG
ACTCTGCGGAGTGTTGAATCTGTTCATCTGTCTGATTTTATCCAGAT
TCGACAATACCTGTCCTCTCAGAGAAACAGGGGCACAGACAGTAATGGAGTAAACCTCTTACAGGAGGCT
CGTCTTCATACAGCACACCTAATCATGATGTATTTACAGTAAACTAGCACATATTACAGCCTACACTGCT
GGAACACACGTCAACAGCTGGACCAACCGGTTTATACACTGAGAGAAAGAGAAAGTGTGAAAATGAGAAG
ACTCTGCGGAGTGTTGAATCTGTTCATCTGTCTGATTTTATCCAGAT