ZFLNCG05503
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 1199.05 |
| ERR023145 | heart | normal | 258.15 |
| SRR592702 | pineal gland | normal | 203.24 |
| SRR592703 | pineal gland | normal | 201.84 |
| SRR372802 | 5 dpf | normal | 184.79 |
| SRR957180 | heart | 7 days after heart tip amputation | 178.89 |
| SRR516127 | skin | male and 5 month | 143.84 |
| ERR023144 | brain | normal | 137.41 |
| SRR592701 | pineal gland | normal | 124.86 |
| SRR516128 | skin | male and 5 month | 121.69 |
Express in tissues
Correlated coding gene
Download
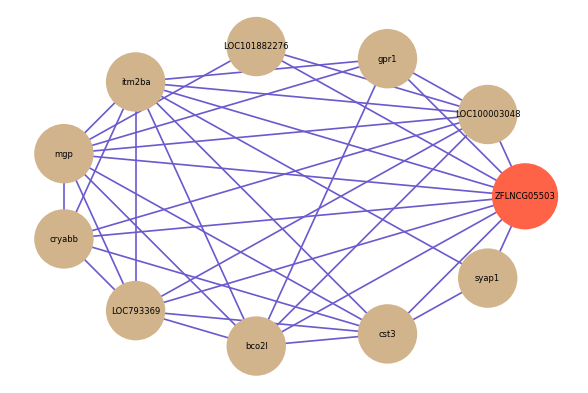
| gene | correlation coefficent |
|---|---|
| LOC100003048 | 0.60 |
| cst3 | 0.59 |
| gpr1 | 0.59 |
| syap1 | 0.58 |
| LOC101882276 | 0.56 |
| itm2ba | 0.56 |
| mgp | 0.55 |
| cryabb | 0.55 |
| LOC793369 | 0.55 |
| bco2l | 0.54 |
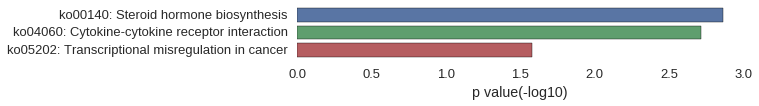
Gene Ontology
Download
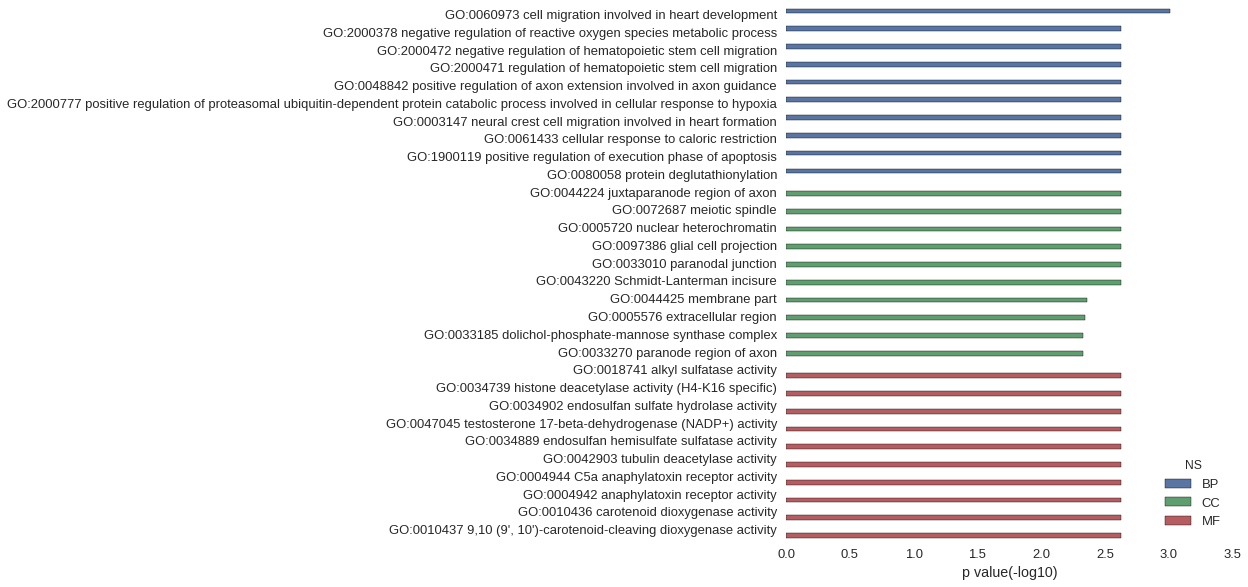
| GO | P value |
|---|---|
| GO:0060973 | 9.87e-04 |
| GO:2000378 | 2.35e-03 |
| GO:2000472 | 2.35e-03 |
| GO:2000471 | 2.35e-03 |
| GO:0048842 | 2.35e-03 |
| GO:2000777 | 2.35e-03 |
| GO:0003147 | 2.35e-03 |
| GO:0061433 | 2.35e-03 |
| GO:1900119 | 2.35e-03 |
| GO:0080058 | 2.35e-03 |
| GO:0044224 | 2.35e-03 |
| GO:0072687 | 2.35e-03 |
| GO:0005720 | 2.35e-03 |
| GO:0097386 | 2.35e-03 |
| GO:0033010 | 2.35e-03 |
| GO:0043220 | 2.35e-03 |
| GO:0044425 | 4.37e-03 |
| GO:0005576 | 4.54e-03 |
| GO:0033185 | 4.68e-03 |
| GO:0033270 | 4.68e-03 |
| GO:0018741 | 2.35e-03 |
| GO:0034739 | 2.35e-03 |
| GO:0034902 | 2.35e-03 |
| GO:0047045 | 2.35e-03 |
| GO:0034889 | 2.35e-03 |
| GO:0042903 | 2.35e-03 |
| GO:0004944 | 2.35e-03 |
| GO:0004942 | 2.35e-03 |
| GO:0010436 | 2.35e-03 |
| GO:0010437 | 2.35e-03 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05503
TTGAATTTCAAGTCATGTGAAGCCATACGATGTCTTTATATGAAGAACGGAGTGATTTATGAGTCTCTAA
AAGCTGAAAATGGCTTGCTAATTTCCTCTAATTAAAAAAAAAATAGCTGAAGTATATTAGCATGATGCAA
TTAGTTACGAGACACAAAGAGAGCCAATAGCATTCGACAACTGAAGAGGATGTAAGACGCTGGC
TTGAATTTCAAGTCATGTGAAGCCATACGATGTCTTTATATGAAGAACGGAGTGATTTATGAGTCTCTAA
AAGCTGAAAATGGCTTGCTAATTTCCTCTAATTAAAAAAAAAATAGCTGAAGTATATTAGCATGATGCAA
TTAGTTACGAGACACAAAGAGAGCCAATAGCATTCGACAACTGAAGAGGATGTAAGACGCTGGC