ZFLNCG05583
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR941753 | posterior pectoral fin | normal | 40.30 |
| SRR800045 | muscle | normal | 8.41 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 8.05 |
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 7.26 |
| SRR941749 | anterior pectoral fin | normal | 5.47 |
| SRR726542 | 5 dpf | infection with control | 4.59 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 4.28 |
| SRR527832 | 16-36 hpf | normal | 2.68 |
| SRR038625 | embryo | traf6 morpholino | 2.41 |
| SRR592698 | pineal gland | normal | 2.23 |
Express in tissues
Correlated coding gene
Download
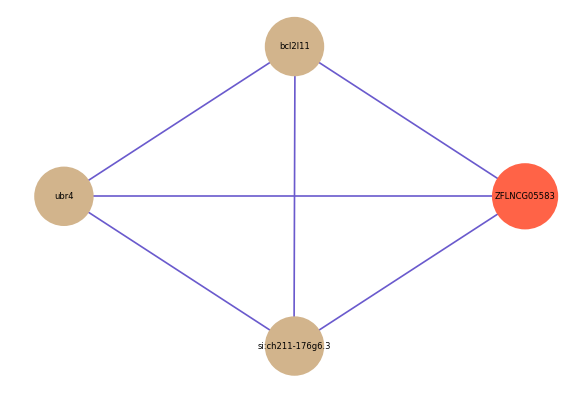
| gene | correlation coefficent |
|---|---|
| bcl2l11 | 0.52 |
| ubr4 | 0.51 |
| si:ch211-176g6.3 | 0.51 |
Gene Ontology
Download
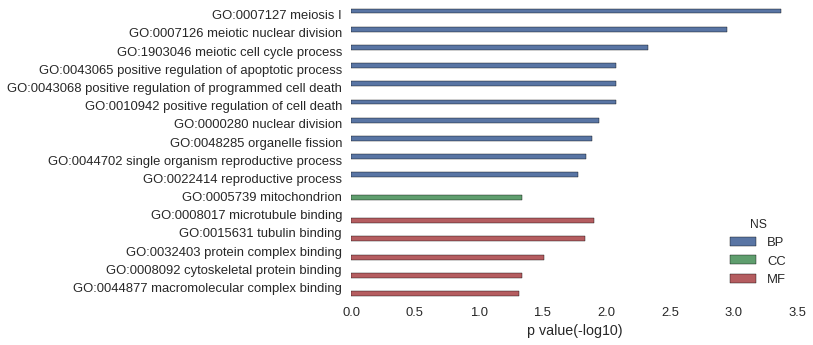
| GO | P value |
|---|---|
| GO:0007127 | 4.26e-04 |
| GO:0007126 | 1.14e-03 |
| GO:1903046 | 4.68e-03 |
| GO:0043065 | 8.37e-03 |
| GO:0043068 | 8.37e-03 |
| GO:0010942 | 8.37e-03 |
| GO:0000280 | 1.13e-02 |
| GO:0048285 | 1.29e-02 |
| GO:0044702 | 1.44e-02 |
| GO:0022414 | 1.66e-02 |
| GO:0005739 | 4.55e-02 |
| GO:0008017 | 1.25e-02 |
| GO:0015631 | 1.47e-02 |
| GO:0032403 | 3.10e-02 |
| GO:0008092 | 4.55e-02 |
| GO:0044877 | 4.86e-02 |
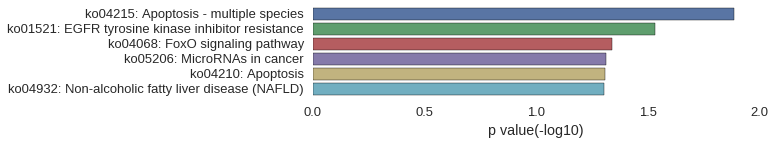
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05583
AGTTGTAGGGGATTTTTCCTCTCTTCTCATCTCCGGTACTGGACCTGGGGCCTTAGGCCCTGCTGGGACA
CCCCTTGCTCTTTCCTCCAACTTGCTTTTGCAGAACCAAGGGAGCATGCTTGCAGCCAGCTTCCAAGAGG
GCCGTAGAGCATCAGATACCTCCCTTACACAGGGTGAGATTTTTACTTCAAAGACACAGTTAATACAATC
AACTCACAGACTTATTATAGACCAAATATCTAAATTGGTTCTGACCAGTCTGTGAAAATTTCTTTAGGTC
TAAAAGCATTCAGACAGCAGCTGAGGAAGAACACTCGTGCGAAAGGCTTACTGGGGTTAAACAAAATCAA
AGGTTTGGCCCGACAAGTGTGCCCACCGTCTTCATGCTCCAGAGGCAGCCGAGGGTCGCTTGGTCCAACT
ATGTGCCCTCCCTCAAGCCTCCAGAGTTCTAGTGGGCCCCGTGAACGCCGTAGCATGTTGGAGGAAGTTC
TGCACCAGCAGAGGTAAGAAAAAATCTGTTCAAAGCAGACATCCAAACACTTAGATACATTTTTGCAATG
TACCTAAACTCTGTTCTTCTTTCATCTTTCAGAATGCTGCAGATTCAGCACCAGCCTCAACCCCAAGTCC
AATTTCTCTCACAGTCTCACCCTTCATCTCCTCCCTCCAACAATCTCTTTGCTCCTGCTGCTCT
AGTTGTAGGGGATTTTTCCTCTCTTCTCATCTCCGGTACTGGACCTGGGGCCTTAGGCCCTGCTGGGACA
CCCCTTGCTCTTTCCTCCAACTTGCTTTTGCAGAACCAAGGGAGCATGCTTGCAGCCAGCTTCCAAGAGG
GCCGTAGAGCATCAGATACCTCCCTTACACAGGGTGAGATTTTTACTTCAAAGACACAGTTAATACAATC
AACTCACAGACTTATTATAGACCAAATATCTAAATTGGTTCTGACCAGTCTGTGAAAATTTCTTTAGGTC
TAAAAGCATTCAGACAGCAGCTGAGGAAGAACACTCGTGCGAAAGGCTTACTGGGGTTAAACAAAATCAA
AGGTTTGGCCCGACAAGTGTGCCCACCGTCTTCATGCTCCAGAGGCAGCCGAGGGTCGCTTGGTCCAACT
ATGTGCCCTCCCTCAAGCCTCCAGAGTTCTAGTGGGCCCCGTGAACGCCGTAGCATGTTGGAGGAAGTTC
TGCACCAGCAGAGGTAAGAAAAAATCTGTTCAAAGCAGACATCCAAACACTTAGATACATTTTTGCAATG
TACCTAAACTCTGTTCTTCTTTCATCTTTCAGAATGCTGCAGATTCAGCACCAGCCTCAACCCCAAGTCC
AATTTCTCTCACAGTCTCACCCTTCATCTCCTCCCTCCAACAATCTCTTTGCTCCTGCTGCTCT