ZFLNCG05635
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR658547 | 6 somite | Gata5/6 morphant | 1.05 |
| SRR658539 | bud | Gata5/6 morphant | 0.96 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 0.70 |
| SRR1708344 | embryo | MUTPS | 0.62 |
| SRR1647683 | spleen | SVCV treatment | 0.60 |
| SRR527840 | 24hpf | normal | 0.55 |
| SRR1035978 | 13 hpf | rx3-/- | 0.54 |
| SRR797911 | embryo | prdm1-/- | 0.54 |
| SRR658535 | bud | Gata5 morphant | 0.54 |
| SRR1708345 | embryo | SIBAS | 0.53 |
Express in tissues
Correlated coding gene
Download
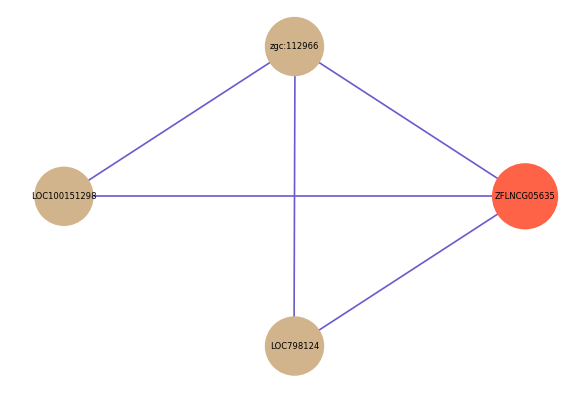
| gene | correlation coefficent |
|---|---|
| zgc:112966 | 0.66 |
| LOC100151298 | 0.58 |
| LOC798124 | 0.54 |
Gene Ontology
Gene Ontology of this lncRNA gene hava does not exist!
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05635
TTATCCACCGTGATGTTTTCAAGGTTTCCACTTAGAAGGTAATTGTAACACCATTCCTGTAAATATCGTG
GAGCTGGACCACCTTGCGCGAGACTTACGGCAAATATTTCCCCAGCAACTCTATAACAATAACGAGAAAA
AAAAACACAATTTCACAAAAATTGTGAAAAACAATTTCTACAGTCAGCAAGTACATTTTCAACTCATTGT
AGGAGCCATTTCTATTCTTTGTTATTTGTGTTGTATGGCACTAGAGAGTGTAATACAACAGCTAAGCTAA
AATAATGTTAAAAAAACACCTTTATTATTTATCTACAGAAAAAAGTACTTAAACTTTGCAGACAAACAGG
ACACTTTGCCTCTGTTGATTAAAAAGCATGTAATGATGTAAAATACACATTAAAAGAAGTAAGAATAATG
TCAACCCAAGCAACTAGGTGATTACCTTTTCTTCAGTAGCAGGAGATAAATCGGGTGAATCAGGAGGTAG
ATTGGACAGCTAGAAATAGACAAAACATTTGACATATTTCCACATGAATCTTACAGAATACTCATTAGGT
TTATTTAAACAATTTATCTTACCTCTGGTTCTGAGTCTGAAAGTGTCTCATATTCAGCAGTG
TTATCCACCGTGATGTTTTCAAGGTTTCCACTTAGAAGGTAATTGTAACACCATTCCTGTAAATATCGTG
GAGCTGGACCACCTTGCGCGAGACTTACGGCAAATATTTCCCCAGCAACTCTATAACAATAACGAGAAAA
AAAAACACAATTTCACAAAAATTGTGAAAAACAATTTCTACAGTCAGCAAGTACATTTTCAACTCATTGT
AGGAGCCATTTCTATTCTTTGTTATTTGTGTTGTATGGCACTAGAGAGTGTAATACAACAGCTAAGCTAA
AATAATGTTAAAAAAACACCTTTATTATTTATCTACAGAAAAAAGTACTTAAACTTTGCAGACAAACAGG
ACACTTTGCCTCTGTTGATTAAAAAGCATGTAATGATGTAAAATACACATTAAAAGAAGTAAGAATAATG
TCAACCCAAGCAACTAGGTGATTACCTTTTCTTCAGTAGCAGGAGATAAATCGGGTGAATCAGGAGGTAG
ATTGGACAGCTAGAAATAGACAAAACATTTGACATATTTCCACATGAATCTTACAGAATACTCATTAGGT
TTATTTAAACAATTTATCTTACCTCTGGTTCTGAGTCTGAAAGTGTCTCATATTCAGCAGTG
