ZFLNCG05681
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR594769 | blood | normal | 14.35 |
| SRR516132 | skin | male and 5 month | 7.26 |
| SRR658543 | 6 somite | Gata5 morphan | 5.80 |
| SRR797908 | embryo | normal | 4.99 |
| SRR594771 | endothelium | normal | 4.72 |
| SRR797914 | embryo | pbx2/4-MO | 4.34 |
| SRR1035978 | 13 hpf | rx3-/- | 4.26 |
| SRR797917 | embryo | pbx2/4-MO and prdm1-/- | 3.79 |
| SRR516133 | skin | male and 5 month | 3.76 |
| SRR797911 | embryo | prdm1-/- | 3.74 |
Express in tissues
Correlated coding gene
Download
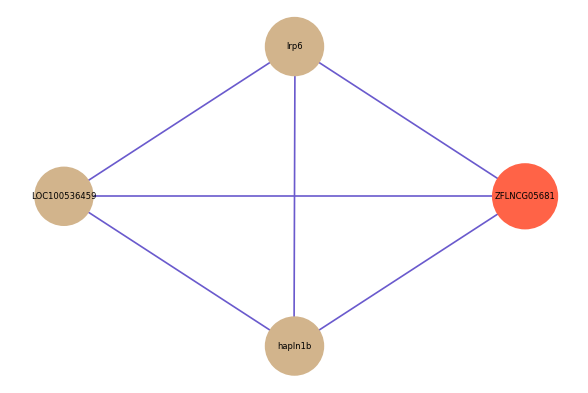
| gene | correlation coefficent |
|---|---|
| lrp6 | 0.52 |
| LOC100536459 | 0.50 |
| hapln1b | 0.50 |
Gene Ontology
Download
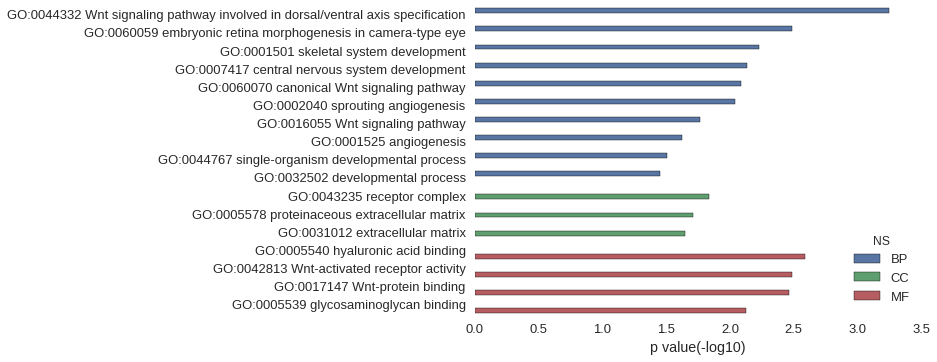
| GO | P value |
|---|---|
| GO:0044332 | 5.68e-04 |
| GO:0060059 | 3.27e-03 |
| GO:0001501 | 5.96e-03 |
| GO:0007417 | 7.38e-03 |
| GO:0060070 | 8.23e-03 |
| GO:0002040 | 9.08e-03 |
| GO:0016055 | 1.70e-02 |
| GO:0001525 | 2.39e-02 |
| GO:0044767 | 3.14e-02 |
| GO:0032502 | 3.56e-02 |
| GO:0043235 | 1.46e-02 |
| GO:0005578 | 1.94e-02 |
| GO:0031012 | 2.23e-02 |
| GO:0005540 | 2.56e-03 |
| GO:0042813 | 3.27e-03 |
| GO:0017147 | 3.41e-03 |
| GO:0005539 | 7.52e-03 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05681
TCCATTCTCAGCCTATGACAAACACACACAAAGAATACAGGTGTGCAACAACATCTCATTTTTACAGAAC
ACAAATGTAAAAGTTGTTGAATTGTAAGCAAAAAAAAATGTCTTTCCCCCCTCACCTTTCTTTTACTTCT
GCCATCTCCAGGTGGTTGGTCAGAGTGGTTCAGATGCTTCATTGATGACTTTTCTTTTGCCTTTCCCGAT
GACACAGATGAAGACGAAGACTGTCTCTCTGTAAGCTCATCTGCACTCGGGACCCTACCCAGAAGACAGA
GGTCTATCCTCACAACTAGTTTTTTCAGGCCTCGATGAGGGATGGTGGGGGACTTTTCAGTCCGGCCAAA
AGGAACGAGTGTATAGAGTTTGTGTCTCCCCCGACTGTGTTTTTCTGTTTCAGGGGTACTTAAGCAGGAC
CTCTCGGGGTGACTACTTGAGCCTGAATTTTGTTTCTCATGCCTGCTCTTTGATACAGTTTTTTTGTTTG
ATGTTGAATCTGCAGGGACTTTCGCAATGTTCCTGGGTGGACTAGACTCAGAGTCTGATTCAGAGGAAGA
TGATGAGGATGAAAAGGAAGATGAGGATGAAGAAGATAATGGATGGGTGGGCTTGACGACAGGAGTAGGG
GTAGGTGGAGAGAGTGGAGGAGAAGGGCTAGAGTCTGTCACATAAGGACGTGAGGAAACGTCTCCGTTAC
TCCACCGTCTCTTCTTCCTCCTCTGCTCCTCAAGGTTGGGTCCATTGCATTCCCTTTGTGTGTGATGCTG
GCTTGTGTGAGGTCTTTGTTTGGGCTGAACTGCCCTCCATTCAGCTTCCGGTTCCC
TCCATTCTCAGCCTATGACAAACACACACAAAGAATACAGGTGTGCAACAACATCTCATTTTTACAGAAC
ACAAATGTAAAAGTTGTTGAATTGTAAGCAAAAAAAAATGTCTTTCCCCCCTCACCTTTCTTTTACTTCT
GCCATCTCCAGGTGGTTGGTCAGAGTGGTTCAGATGCTTCATTGATGACTTTTCTTTTGCCTTTCCCGAT
GACACAGATGAAGACGAAGACTGTCTCTCTGTAAGCTCATCTGCACTCGGGACCCTACCCAGAAGACAGA
GGTCTATCCTCACAACTAGTTTTTTCAGGCCTCGATGAGGGATGGTGGGGGACTTTTCAGTCCGGCCAAA
AGGAACGAGTGTATAGAGTTTGTGTCTCCCCCGACTGTGTTTTTCTGTTTCAGGGGTACTTAAGCAGGAC
CTCTCGGGGTGACTACTTGAGCCTGAATTTTGTTTCTCATGCCTGCTCTTTGATACAGTTTTTTTGTTTG
ATGTTGAATCTGCAGGGACTTTCGCAATGTTCCTGGGTGGACTAGACTCAGAGTCTGATTCAGAGGAAGA
TGATGAGGATGAAAAGGAAGATGAGGATGAAGAAGATAATGGATGGGTGGGCTTGACGACAGGAGTAGGG
GTAGGTGGAGAGAGTGGAGGAGAAGGGCTAGAGTCTGTCACATAAGGACGTGAGGAAACGTCTCCGTTAC
TCCACCGTCTCTTCTTCCTCCTCTGCTCCTCAAGGTTGGGTCCATTGCATTCCCTTTGTGTGTGATGCTG
GCTTGTGTGAGGTCTTTGTTTGGGCTGAACTGCCCTCCATTCAGCTTCCGGTTCCC
