ZFLNCG05781
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR594771 | endothelium | normal | 13.49 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 12.57 |
| SRR1035237 | liver | transgenic UHRF1 | 11.13 |
| SRR1004789 | larvae | impdh2 morpholino | 10.87 |
| SRR527835 | tail | normal | 10.21 |
| SRR658543 | 6 somite | Gata5 morphan | 10.17 |
| SRR527833 | 5 dpf | normal | 9.43 |
| SRR1569491 | embryo | normal | 9.23 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 8.15 |
| SRR527832 | 16-36 hpf | normal | 7.61 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| si:dkey-159f12.2 | 0.50 |
Gene Ontology
Download
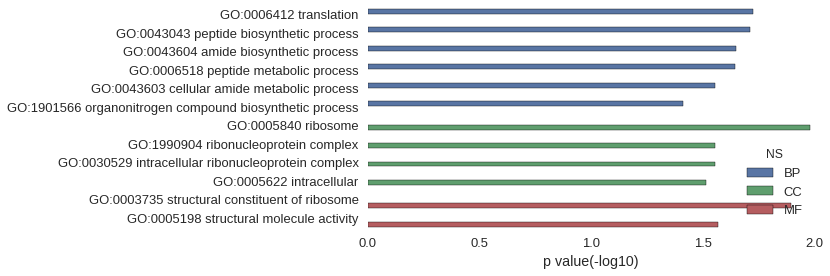
| GO | P value |
|---|---|
| GO:0006412 | 1.88e-02 |
| GO:0043043 | 1.93e-02 |
| GO:0043604 | 2.25e-02 |
| GO:0006518 | 2.27e-02 |
| GO:0043603 | 2.79e-02 |
| GO:1901566 | 3.88e-02 |
| GO:0005840 | 1.04e-02 |
| GO:1990904 | 2.79e-02 |
| GO:0030529 | 2.79e-02 |
| GO:0005622 | 3.06e-02 |
| GO:0003735 | 1.27e-02 |
| GO:0005198 | 2.70e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT08923 | NONHSAT183471; | ENSMUST00000117815; | Direct blastn |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05781
AGCCAAAGGCAGAGGGGTGGGGAGAGAGAACAAGTTTTGTCCGTTGAGCTCCGTCTTGGGAGTGGCGAAT
GTTCCGAGAAAAAGCTTTTATGAAAGAACGGCATTGGATCGGCGGATGCCCACTAGGTTGTCTGCGCTGA
ATGATCTCCTCATGGCCACTCTGCTCAGATCTTTGTACTTATCCTCACACAGTCTCGAGATTGCCTTTGA
AATGAAGAACAAGTTGAATGAGAACGCTTGTATTTGAACTAGACCTTTAATAAAAGTGATTAACTCTTTG
ACTGCCCAATCTACCAAACGGTTGACCATACCAAATATTTGATTTGTCGGTTTATGGTAAAAGTATCAGA
CTTAGTACATATACTATGAAAACATTTTTGCATACTAAAATCACCTTTTTTTTGAAGTATGGGATAAAGT
AATTCAGATTTTCATTAAACATGTTTTCTTGGTTTATTTTTTTAAATGAAACCAAATTAAGTGCAGCAGG
ATTATGTTTGATTTTTTTTATTTTTTTTTTATAAACCAACAATGCTGTGTGTGTAAACCATTATATAAAA
CAGTCATTCTTTAATTGACTTCAACAGTCATTACTTAAAACAGGACATCAGGAGTTAAGGAATGCAATCT
AAATTTTTTCTCTTGGTGGAGTAGTTAAAGGGTTAAAGTAAACAAAAATTAATAAAAGTTTTAAGTGTTT
ACCTCTAATTTCTGAGCTCGTTCATTGCTTAATGGCTCCAGAGGAAAGCTCAGGTTGTTGTCGTCTGCTA
AAGAGCGCAGGTCAAAGTAATACTTGGCGTAGACGCTGGCCGGTACGTTAATGTTAAACTGCAGCAGCTC
TAGAAAATGTCTTTCCATCTCATTCCTGAAAAACAAGATAACAAGTTAGGGAGGAAATATGTCCTTTCTG
ACTTGAAAGTCTAAAGGCAACAGTTTTAGTGTAAATCGAACCTAAACAGATTTGAAAAGTCGACACAACA
GCT
AGCCAAAGGCAGAGGGGTGGGGAGAGAGAACAAGTTTTGTCCGTTGAGCTCCGTCTTGGGAGTGGCGAAT
GTTCCGAGAAAAAGCTTTTATGAAAGAACGGCATTGGATCGGCGGATGCCCACTAGGTTGTCTGCGCTGA
ATGATCTCCTCATGGCCACTCTGCTCAGATCTTTGTACTTATCCTCACACAGTCTCGAGATTGCCTTTGA
AATGAAGAACAAGTTGAATGAGAACGCTTGTATTTGAACTAGACCTTTAATAAAAGTGATTAACTCTTTG
ACTGCCCAATCTACCAAACGGTTGACCATACCAAATATTTGATTTGTCGGTTTATGGTAAAAGTATCAGA
CTTAGTACATATACTATGAAAACATTTTTGCATACTAAAATCACCTTTTTTTTGAAGTATGGGATAAAGT
AATTCAGATTTTCATTAAACATGTTTTCTTGGTTTATTTTTTTAAATGAAACCAAATTAAGTGCAGCAGG
ATTATGTTTGATTTTTTTTATTTTTTTTTTATAAACCAACAATGCTGTGTGTGTAAACCATTATATAAAA
CAGTCATTCTTTAATTGACTTCAACAGTCATTACTTAAAACAGGACATCAGGAGTTAAGGAATGCAATCT
AAATTTTTTCTCTTGGTGGAGTAGTTAAAGGGTTAAAGTAAACAAAAATTAATAAAAGTTTTAAGTGTTT
ACCTCTAATTTCTGAGCTCGTTCATTGCTTAATGGCTCCAGAGGAAAGCTCAGGTTGTTGTCGTCTGCTA
AAGAGCGCAGGTCAAAGTAATACTTGGCGTAGACGCTGGCCGGTACGTTAATGTTAAACTGCAGCAGCTC
TAGAAAATGTCTTTCCATCTCATTCCTGAAAAACAAGATAACAAGTTAGGGAGGAAATATGTCCTTTCTG
ACTTGAAAGTCTAAAGGCAACAGTTTTAGTGTAAATCGAACCTAAACAGATTTGAAAAGTCGACACAACA
GCT
