ZFLNCG05846
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 8.56 |
| ERR594416 | retina | transgenic flk1 | 6.41 |
| SRR592702 | pineal gland | normal | 6.23 |
| SRR038627 | embryo | control morpholino | 6.22 |
| SRR1291417 | 5 dpi | injected marinum | 6.21 |
| ERR023144 | brain | normal | 5.90 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 5.79 |
| ERR023143 | swim bladder | normal | 5.67 |
| ERR594417 | retina | normal | 5.61 |
| SRR1291414 | 5 dpi | normal | 5.37 |
Express in tissues
Correlated coding gene
Download
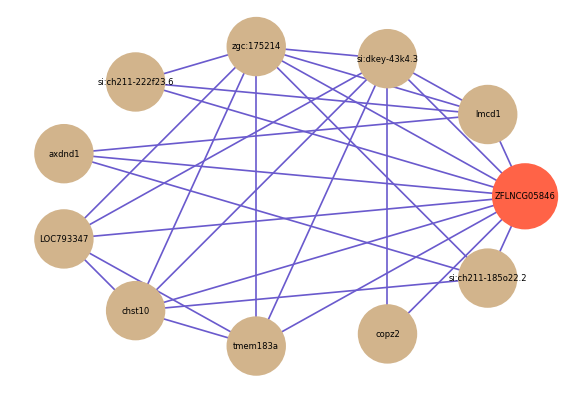
| gene | correlation coefficent |
|---|---|
| lmcd1 | 0.55 |
| LOC793347 | 0.53 |
| copz2 | 0.53 |
| si:ch211-185o22.2 | 0.53 |
| si:dkey-43k4.3 | 0.53 |
| zgc:175214 | 0.51 |
| si:ch211-222f23.6 | 0.51 |
| chst10 | 0.51 |
| tmem183a | 0.50 |
| axdnd1 | 0.50 |
Gene Ontology
Download
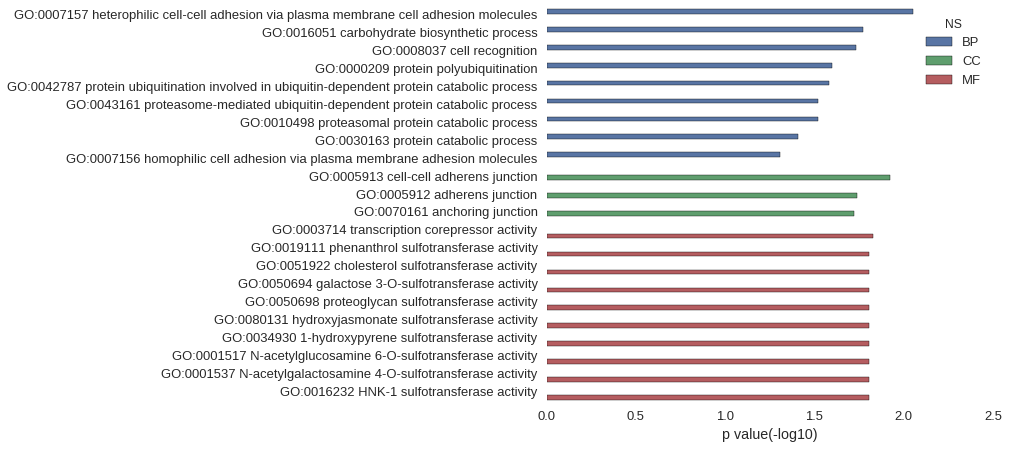
| GO | P value |
|---|---|
| GO:0007157 | 8.92e-03 |
| GO:0016051 | 1.69e-02 |
| GO:0008037 | 1.86e-02 |
| GO:0000209 | 2.53e-02 |
| GO:0042787 | 2.62e-02 |
| GO:0043161 | 3.03e-02 |
| GO:0010498 | 3.03e-02 |
| GO:0030163 | 3.90e-02 |
| GO:0007156 | 4.93e-02 |
| GO:0005913 | 1.19e-02 |
| GO:0005912 | 1.82e-02 |
| GO:0070161 | 1.90e-02 |
| GO:0003714 | 1.48e-02 |
| GO:0019111 | 1.57e-02 |
| GO:0051922 | 1.57e-02 |
| GO:0050694 | 1.57e-02 |
| GO:0050698 | 1.57e-02 |
| GO:0080131 | 1.57e-02 |
| GO:0034930 | 1.57e-02 |
| GO:0001517 | 1.57e-02 |
| GO:0001537 | 1.57e-02 |
| GO:0016232 | 1.57e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05846
GACTGGTCTTTGCAGGAAAACCACTTGCTTAGTGGCCAAATTCCCTTCACTGCAGAAACAGAATAAAAAA
AAAACACTCAGCTAGCCATATAATAACAGATTGTAAAATAAACTTGATTAAACAGCTGTGAGGATCGCAC
CTGTCATGGCGAATAATCGGGGTGGGCAAAACACAGGTGAGCAGCCAGCCAAACTCTGCTAACGTGTGCA
GCAGAGGCCCATAGTCTGTCTTCACATTTGAATCCTAAAACACAAACCCATTAGCGACATCATCAAAATG
AGTTTAACACCATGCTAGATAATGACAGCTTCCACACATTCAATCACACCTCAATAACAGTCCACTGTTC
CACTACAATAGTGTCATTGGCAGGTGGCGCTGTGCTTTTCTCTTCATAGACAAACAGACCCTCTAGAGAC
CTGGGCACTGGCTCTCCTGCCAAAACAACAATATAACATCTAGATTAGGTGATAAGAACTCATAAAGCCA
CAGCTCATTGCTGGATTTTTGCATAATAGGTGCATGTGTATGACCTAGATATTCATTTAAAGAGACAGTA
CCCACACACACAGACAAAAAAAAGACAATTACGTTCGTTAATGGGATTCTTTTCTTTGTTGAACATGAAT
GAGGATATTTTGAAGAATGTTCAAACAGGAGCCATTACTAGTAATGAAGTCAGTAGTTAATCTGTG
GACTGGTCTTTGCAGGAAAACCACTTGCTTAGTGGCCAAATTCCCTTCACTGCAGAAACAGAATAAAAAA
AAAACACTCAGCTAGCCATATAATAACAGATTGTAAAATAAACTTGATTAAACAGCTGTGAGGATCGCAC
CTGTCATGGCGAATAATCGGGGTGGGCAAAACACAGGTGAGCAGCCAGCCAAACTCTGCTAACGTGTGCA
GCAGAGGCCCATAGTCTGTCTTCACATTTGAATCCTAAAACACAAACCCATTAGCGACATCATCAAAATG
AGTTTAACACCATGCTAGATAATGACAGCTTCCACACATTCAATCACACCTCAATAACAGTCCACTGTTC
CACTACAATAGTGTCATTGGCAGGTGGCGCTGTGCTTTTCTCTTCATAGACAAACAGACCCTCTAGAGAC
CTGGGCACTGGCTCTCCTGCCAAAACAACAATATAACATCTAGATTAGGTGATAAGAACTCATAAAGCCA
CAGCTCATTGCTGGATTTTTGCATAATAGGTGCATGTGTATGACCTAGATATTCATTTAAAGAGACAGTA
CCCACACACACAGACAAAAAAAAGACAATTACGTTCGTTAATGGGATTCTTTTCTTTGTTGAACATGAAT
GAGGATATTTTGAAGAATGTTCAAACAGGAGCCATTACTAGTAATGAAGTCAGTAGTTAATCTGTG

