ZFLNCG06069
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR519752 | 7 dpf | VD3 treatment | 25.79 |
| SRR519722 | 4 dpf | FETOH treatment | 23.42 |
| SRR519742 | 4 dpf | VD3 treatment | 22.92 |
| SRR519727 | 6 dpf | FETOH treatment | 20.48 |
| SRR519747 | 6 dpf | VD3 treatment | 18.31 |
| SRR519717 | 2 dpf | FETOH treatment | 17.65 |
| SRR519737 | 2 dpf | VD3 treatment | 17.54 |
| SRR519732 | 7 dpf | FETOH treatment | 15.52 |
| SRR1648855 | brain | normal | 14.46 |
| SRR1648856 | brain | normal | 12.91 |
Express in tissues
Correlated coding gene
Download
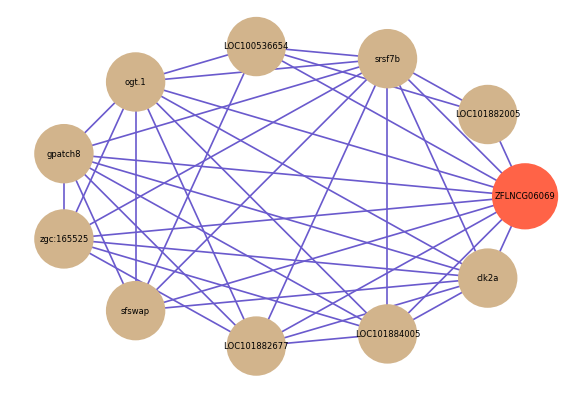
| gene | correlation coefficent |
|---|---|
| LOC101882005 | 0.67 |
| ogt.1 | 0.66 |
| gpatch8 | 0.63 |
| srsf7b | 0.62 |
| zgc:165525 | 0.61 |
| sfswap | 0.61 |
| LOC101882677 | 0.60 |
| LOC101884005 | 0.59 |
| LOC100536654 | 0.58 |
| clk2a | 0.58 |
Gene Ontology
Download
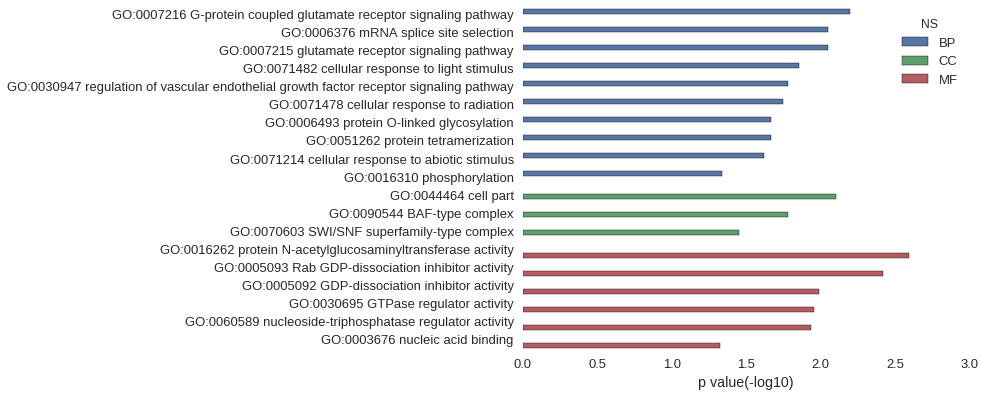
| GO | P value |
|---|---|
| GO:0007216 | 6.38e-03 |
| GO:0006376 | 8.92e-03 |
| GO:0007215 | 8.92e-03 |
| GO:0071482 | 1.40e-02 |
| GO:0030947 | 1.65e-02 |
| GO:0071478 | 1.78e-02 |
| GO:0006493 | 2.15e-02 |
| GO:0051262 | 2.15e-02 |
| GO:0071214 | 2.40e-02 |
| GO:0016310 | 4.59e-02 |
| GO:0044464 | 7.86e-03 |
| GO:0090544 | 1.65e-02 |
| GO:0070603 | 3.52e-02 |
| GO:0016262 | 2.56e-03 |
| GO:0005093 | 3.83e-03 |
| GO:0005092 | 1.02e-02 |
| GO:0030695 | 1.11e-02 |
| GO:0060589 | 1.16e-02 |
| GO:0003676 | 4.75e-02 |
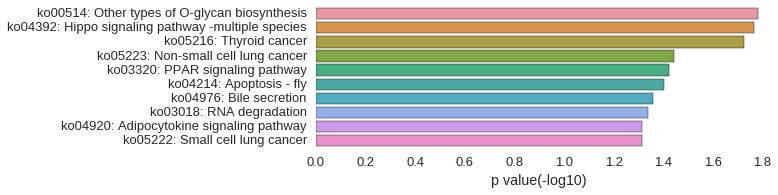
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06069
CACACACCGATGGCTCCAGGAAATCCTGCGGTACACACACACACACACACACACACACACACACACACTC
AAGGTGAATGGGAACAGCAGAAACAGGTCTCTGTGTGTGTGTGTGTGTGTGTGTGTGTGCCGATGAGGCA
CTGACGGTGTTGAGGAAGCGCAGTGAGCGGATGGAGAGATCAGTCCTGCTGAACAGACGAGTGCAGTCGC
TGTAGTCACCCTCCTCCAACACACACTGATGACCTCTGAACCCTGCAGCGGAGAACACCACCCATCTGAC
ACACACACACACACACACACACACACACACACACACGCGCACACACGCGCACACATACACACACGCATTA
CTGAAGTGTTCTCAGTGGTTGGTCAGTGTTGTACAGGTGTGTGTTGTTCCTCCTCAGACACACACATTCC
TGCAGTGACCCTCAGGGACCCCGGGGCCTCCAGCTCCTCCAGACAGGCCCTGTGCTCCAGCACTGCCCTG
CACTGACCCTCCAGCAGCGGACGCGCAAACAACA
CACACACCGATGGCTCCAGGAAATCCTGCGGTACACACACACACACACACACACACACACACACACACTC
AAGGTGAATGGGAACAGCAGAAACAGGTCTCTGTGTGTGTGTGTGTGTGTGTGTGTGTGCCGATGAGGCA
CTGACGGTGTTGAGGAAGCGCAGTGAGCGGATGGAGAGATCAGTCCTGCTGAACAGACGAGTGCAGTCGC
TGTAGTCACCCTCCTCCAACACACACTGATGACCTCTGAACCCTGCAGCGGAGAACACCACCCATCTGAC
ACACACACACACACACACACACACACACACACACACGCGCACACACGCGCACACATACACACACGCATTA
CTGAAGTGTTCTCAGTGGTTGGTCAGTGTTGTACAGGTGTGTGTTGTTCCTCCTCAGACACACACATTCC
TGCAGTGACCCTCAGGGACCCCGGGGCCTCCAGCTCCTCCAGACAGGCCCTGTGCTCCAGCACTGCCCTG
CACTGACCCTCCAGCAGCGGACGCGCAAACAACA