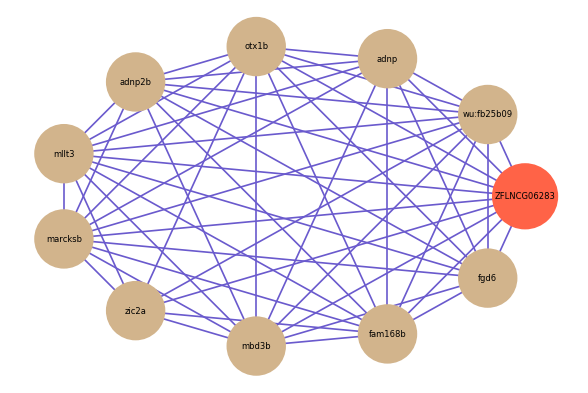
ZFLNCG06283
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR594771 | endothelium | normal | 30.62 |
| SRR658547 | 6 somite | Gata5/6 morphant | 30.21 |
| SRR658541 | 6 somite | normal | 27.48 |
| SRR749515 | 24 hpf | eif3ha morphant | 25.05 |
| SRR658545 | 6 somite | Gata6 morphant | 24.70 |
| ERR023144 | brain | normal | 22.95 |
| SRR527836 | 24hpf | normal | 22.51 |
| SRR372796 | bud | normal | 21.49 |
| SRR658537 | bud | Gata6 morphant | 20.95 |
| SRR658543 | 6 somite | Gata5 morphan | 20.78 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
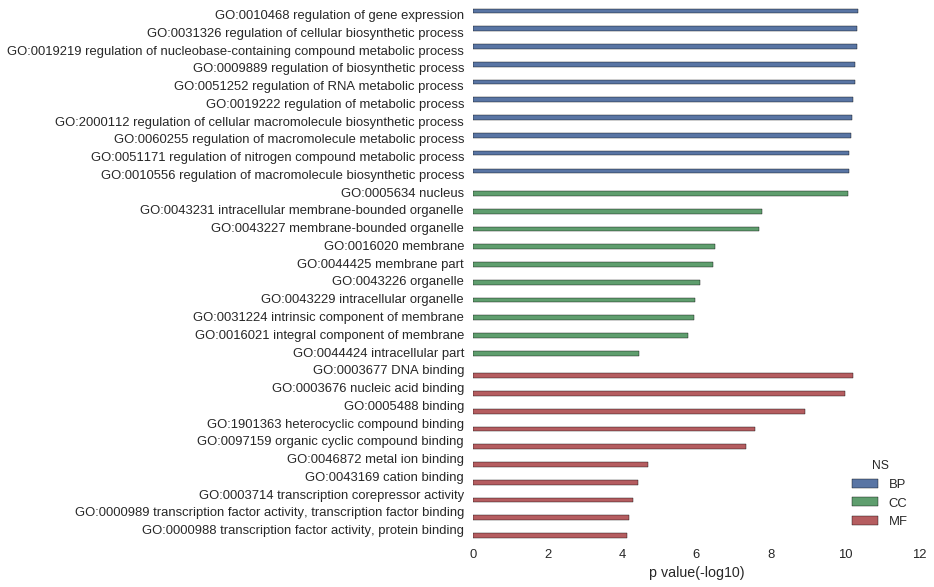
| GO | P value |
|---|---|
| GO:0010468 | 4.56e-11 |
| GO:0031326 | 4.77e-11 |
| GO:0019219 | 4.95e-11 |
| GO:0009889 | 5.36e-11 |
| GO:0051252 | 5.58e-11 |
| GO:0019222 | 6.29e-11 |
| GO:2000112 | 6.41e-11 |
| GO:0060255 | 7.04e-11 |
| GO:0051171 | 8.09e-11 |
| GO:0010556 | 8.09e-11 |
| GO:0005634 | 8.16e-11 |
| GO:0043231 | 1.76e-08 |
| GO:0043227 | 2.04e-08 |
| GO:0016020 | 3.23e-07 |
| GO:0044425 | 3.47e-07 |
| GO:0043226 | 8.11e-07 |
| GO:0043229 | 1.11e-06 |
| GO:0031224 | 1.16e-06 |
| GO:0016021 | 1.68e-06 |
| GO:0044424 | 3.51e-05 |
| GO:0003677 | 6.04e-11 |
| GO:0003676 | 1.04e-10 |
| GO:0005488 | 1.23e-09 |
| GO:1901363 | 2.66e-08 |
| GO:0097159 | 4.77e-08 |
| GO:0046872 | 2.01e-05 |
| GO:0043169 | 3.76e-05 |
| GO:0003714 | 4.95e-05 |
| GO:0000989 | 6.50e-05 |
| GO:0000988 | 7.20e-05 |
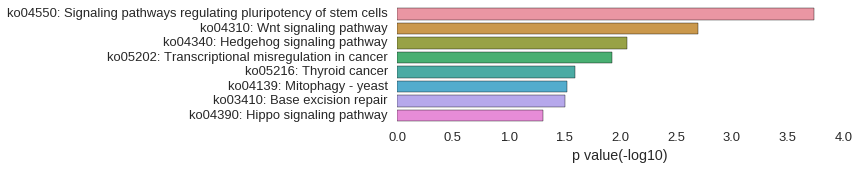
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06283
CGAATACAAAACTAAATTGAAATTGTGGTTTTTGAGAGGAAGGCAGGTCACAATTGAGTAAAAATGACAG
AATTATTAGTTATAATCTCAGCAATATGATGAATTGTGAAAAGGTTAATAAACTTACAAGCCTGAAAACA
ATTGAAATTTGAGTTCCTTCCAACTTTCGGTTAAATTCAGTTACCTCTCCTCAAACTGAATCAGAGTTGA
CAGTCAGATCTGCATAACAACCTCAGAACTGTAATTTAATTTGGCAGATTTATTGCATTACTTTTTGATT
ACAATTCTGAGGTTGTTATGTAGATCTGACTGTCAATTCTGATTACATTTGCGGTGAGTTAAGTGAAGTT
AACCCAAAGTTGGAAAGAACTCAAATTTGAATTATTTTCAGGGTTGTAACTT
CGAATACAAAACTAAATTGAAATTGTGGTTTTTGAGAGGAAGGCAGGTCACAATTGAGTAAAAATGACAG
AATTATTAGTTATAATCTCAGCAATATGATGAATTGTGAAAAGGTTAATAAACTTACAAGCCTGAAAACA
ATTGAAATTTGAGTTCCTTCCAACTTTCGGTTAAATTCAGTTACCTCTCCTCAAACTGAATCAGAGTTGA
CAGTCAGATCTGCATAACAACCTCAGAACTGTAATTTAATTTGGCAGATTTATTGCATTACTTTTTGATT
ACAATTCTGAGGTTGTTATGTAGATCTGACTGTCAATTCTGATTACATTTGCGGTGAGTTAAGTGAAGTT
AACCCAAAGTTGGAAAGAACTCAAATTTGAATTATTTTCAGGGTTGTAACTT