ZFLNCG06305
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR891512 | blood | normal | 106.43 |
| SRR516133 | skin | male and 5 month | 50.41 |
| SRR941753 | posterior pectoral fin | normal | 29.92 |
| SRR516124 | skin | male and 3.5 year | 29.81 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 29.50 |
| SRR516121 | skin | male and 3.5 year | 23.03 |
| SRR372802 | 5 dpf | normal | 22.31 |
| SRR516123 | skin | male and 3.5 year | 18.80 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 17.79 |
| SRR516134 | skin | male and 5 month | 16.83 |
Express in tissues
Correlated coding gene
Download
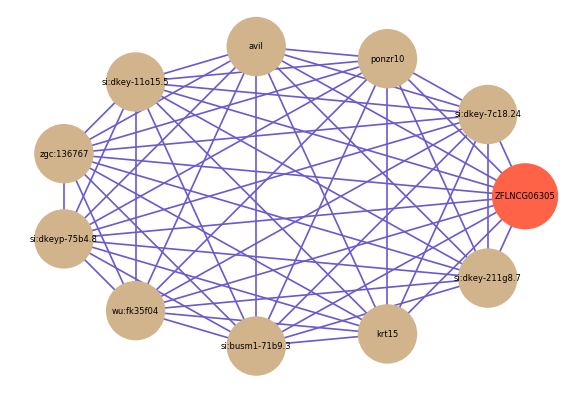
| gene | correlation coefficent |
|---|---|
| si:dkey-7c18.24 | 0.66 |
| ponzr10 | 0.66 |
| avil | 0.62 |
| si:dkey-11o15.5 | 0.62 |
| zgc:136767 | 0.61 |
| si:dkeyp-75b4.8 | 0.61 |
| wu:fk35f04 | 0.61 |
| si:busm1-71b9.3 | 0.61 |
| krt15 | 0.60 |
| si:dkey-211g8.7 | 0.60 |
Gene Ontology
Download
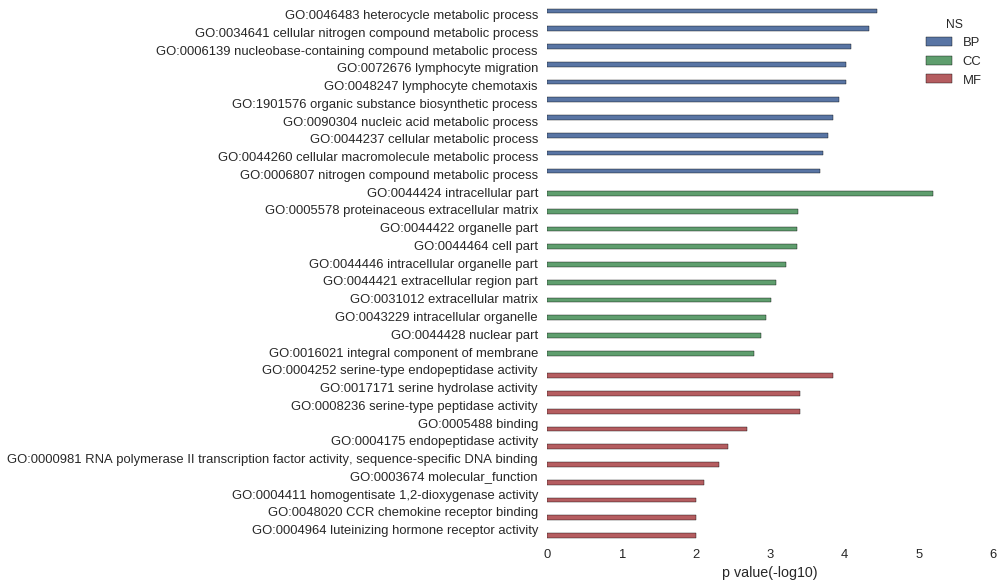
| GO | P value |
|---|---|
| GO:0046483 | 3.63e-05 |
| GO:0034641 | 4.69e-05 |
| GO:0006139 | 8.10e-05 |
| GO:0072676 | 9.69e-05 |
| GO:0048247 | 9.69e-05 |
| GO:1901576 | 1.18e-04 |
| GO:0090304 | 1.43e-04 |
| GO:0044237 | 1.66e-04 |
| GO:0044260 | 1.94e-04 |
| GO:0006807 | 2.12e-04 |
| GO:0044424 | 6.56e-06 |
| GO:0005578 | 4.17e-04 |
| GO:0044422 | 4.37e-04 |
| GO:0044464 | 4.39e-04 |
| GO:0044446 | 6.23e-04 |
| GO:0044421 | 8.42e-04 |
| GO:0031012 | 9.74e-04 |
| GO:0043229 | 1.13e-03 |
| GO:0044428 | 1.34e-03 |
| GO:0016021 | 1.65e-03 |
| GO:0004252 | 1.42e-04 |
| GO:0017171 | 3.99e-04 |
| GO:0008236 | 3.99e-04 |
| GO:0005488 | 2.06e-03 |
| GO:0004175 | 3.66e-03 |
| GO:0000981 | 4.81e-03 |
| GO:0003674 | 7.88e-03 |
| GO:0004411 | 9.88e-03 |
| GO:0048020 | 9.88e-03 |
| GO:0004964 | 9.88e-03 |
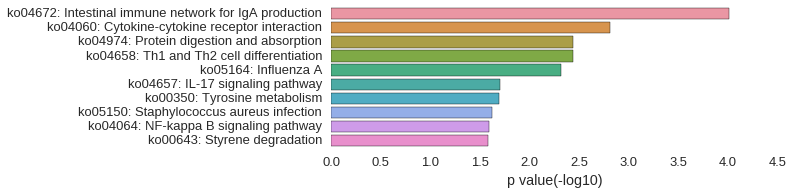
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06305
GACTGAGTGCAGAGGATGTTTATTGACAAATGTGACAATGTAATAGGGGTTAGGTGCAGAACAGTTCAAT
GGTGAATGTATTGGGTAGTGCTGGTGAATCCCGGATAATGCAGCCAGAATCCAAGTAATGCCGGAAGGGT
AGGAGACTGAGCAGTGGGCAGGGTTGGACAAAAACAAAGAACAGAGACAAGGCACACAGAACAGACAGGG
GGAACAAAACAAAAC
GACTGAGTGCAGAGGATGTTTATTGACAAATGTGACAATGTAATAGGGGTTAGGTGCAGAACAGTTCAAT
GGTGAATGTATTGGGTAGTGCTGGTGAATCCCGGATAATGCAGCCAGAATCCAAGTAATGCCGGAAGGGT
AGGAGACTGAGCAGTGGGCAGGGTTGGACAAAAACAAAGAACAGAGACAAGGCACACAGAACAGACAGGG
GGAACAAAACAAAAC