ZFLNCG06337
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 504.38 |
| ERR023143 | swim bladder | normal | 312.30 |
| SRR516131 | skin | male and 5 month | 115.66 |
| ERR023146 | head kidney | normal | 90.40 |
| SRR516134 | skin | male and 5 month | 86.67 |
| SRR516122 | skin | male and 3.5 year | 65.20 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 60.12 |
| SRR516133 | skin | male and 5 month | 59.39 |
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 58.37 |
| SRR1028004 | head | normal | 53.65 |
Express in tissues
Correlated coding gene
Download
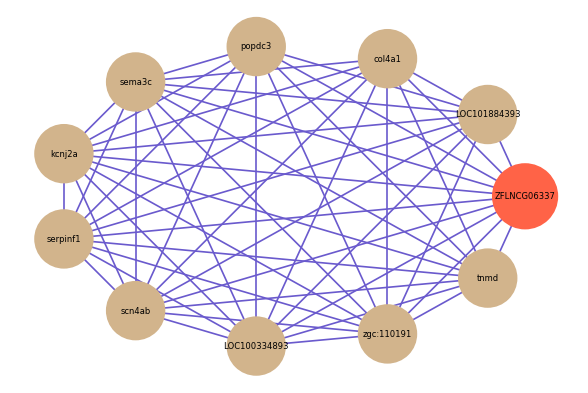
| gene | correlation coefficent |
|---|---|
| LOC101884393 | 0.66 |
| tnmd | 0.65 |
| col4a1 | 0.65 |
| popdc3 | 0.65 |
| sema3c | 0.64 |
| kcnj2a | 0.64 |
| serpinf1 | 0.64 |
| scn4ab | 0.63 |
| LOC100334893 | 0.63 |
| zgc:110191 | 0.63 |
Gene Ontology
Download
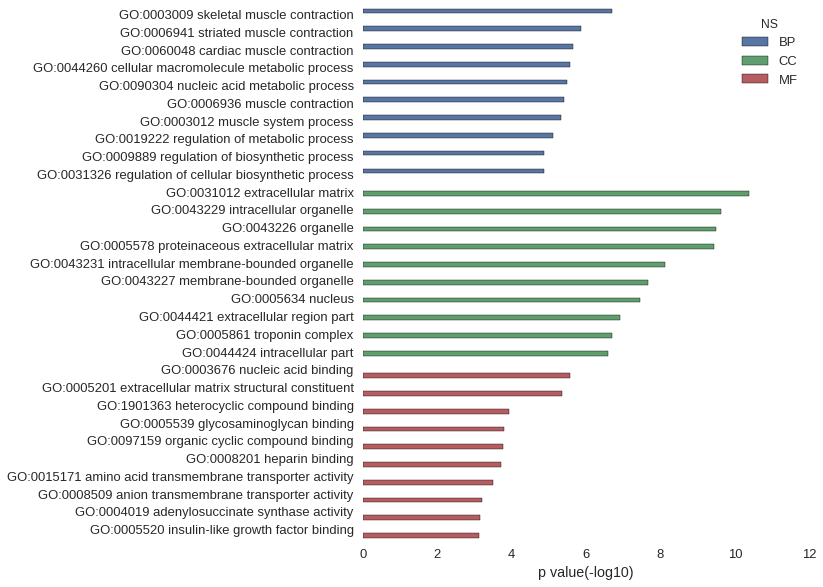
| GO | P value |
|---|---|
| GO:0003009 | 2.05e-07 |
| GO:0006941 | 1.38e-06 |
| GO:0060048 | 2.26e-06 |
| GO:0044260 | 2.69e-06 |
| GO:0090304 | 3.26e-06 |
| GO:0006936 | 3.90e-06 |
| GO:0003012 | 4.58e-06 |
| GO:0019222 | 7.61e-06 |
| GO:0009889 | 1.35e-05 |
| GO:0031326 | 1.35e-05 |
| GO:0031012 | 4.16e-11 |
| GO:0043229 | 2.36e-10 |
| GO:0043226 | 3.12e-10 |
| GO:0005578 | 3.59e-10 |
| GO:0043231 | 7.41e-09 |
| GO:0043227 | 2.14e-08 |
| GO:0005634 | 3.48e-08 |
| GO:0044421 | 1.24e-07 |
| GO:0005861 | 2.05e-07 |
| GO:0044424 | 2.51e-07 |
| GO:0003676 | 2.65e-06 |
| GO:0005201 | 4.55e-06 |
| GO:1901363 | 1.15e-04 |
| GO:0005539 | 1.61e-04 |
| GO:0097159 | 1.66e-04 |
| GO:0008201 | 1.89e-04 |
| GO:0015171 | 3.22e-04 |
| GO:0008509 | 6.21e-04 |
| GO:0004019 | 7.09e-04 |
| GO:0005520 | 7.43e-04 |
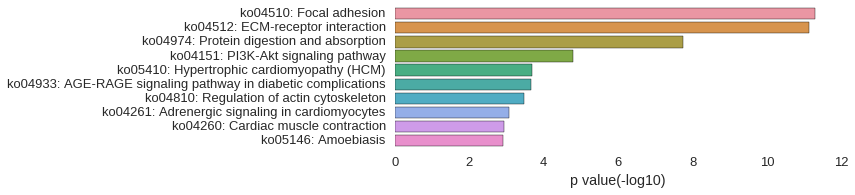
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06337
TTTTATTTTCAAAATCCTTTCATTAATACTGTATAAAATTCAAAGAATGGCATTTATTAGAAAGATACTG
CTTTTTGTAACATTATTAATTGCTTTTAGCAATCTCATTCATCCTGGCTGAATAAAAGTATTACTTAAAT
AATATATTCTTTTATTACTGACCCCAGAACTTTTCAAGCACCACAAACAGCATATTGGAGTGATTTCTGA
ATGACCAGACTCAATAATGAC
TTTTATTTTCAAAATCCTTTCATTAATACTGTATAAAATTCAAAGAATGGCATTTATTAGAAAGATACTG
CTTTTTGTAACATTATTAATTGCTTTTAGCAATCTCATTCATCCTGGCTGAATAAAAGTATTACTTAAAT
AATATATTCTTTTATTACTGACCCCAGAACTTTTCAAGCACCACAAACAGCATATTGGAGTGATTTCTGA
ATGACCAGACTCAATAATGAC