ZFLNCG06338
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 401.66 |
| ERR023145 | heart | normal | 298.73 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 232.01 |
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 225.07 |
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 166.21 |
| SRR516131 | skin | male and 5 month | 129.00 |
| SRR516122 | skin | male and 3.5 year | 119.26 |
| SRR516133 | skin | male and 5 month | 104.66 |
| SRR1028002 | head | normal | 91.88 |
| ERR023146 | head kidney | normal | 88.78 |
Express in tissues
Correlated coding gene
Download
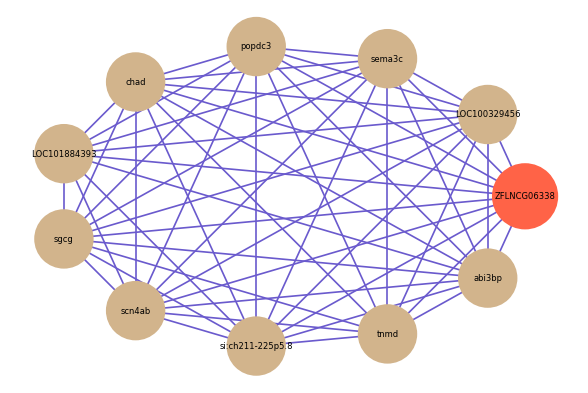
| gene | correlation coefficent |
|---|---|
| LOC100329456 | 0.68 |
| sema3c | 0.66 |
| popdc3 | 0.66 |
| chad | 0.66 |
| LOC101884393 | 0.65 |
| sgcg | 0.65 |
| scn4ab | 0.65 |
| si:ch211-225p5.8 | 0.64 |
| tnmd | 0.64 |
| abi3bp | 0.64 |
Gene Ontology
Download
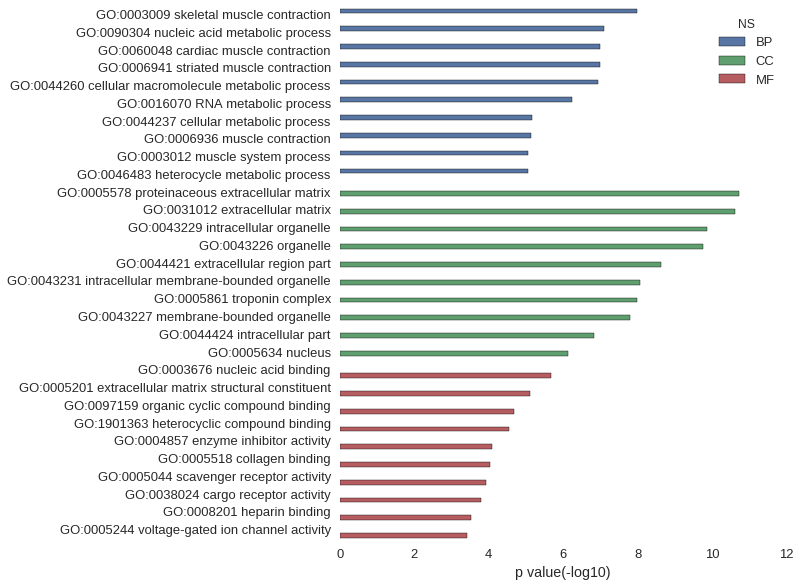
| GO | P value |
|---|---|
| GO:0003009 | 1.04e-08 |
| GO:0090304 | 7.82e-08 |
| GO:0060048 | 1.02e-07 |
| GO:0006941 | 1.04e-07 |
| GO:0044260 | 1.13e-07 |
| GO:0016070 | 5.82e-07 |
| GO:0044237 | 7.08e-06 |
| GO:0006936 | 7.34e-06 |
| GO:0003012 | 8.61e-06 |
| GO:0046483 | 8.65e-06 |
| GO:0005578 | 1.83e-11 |
| GO:0031012 | 2.41e-11 |
| GO:0043229 | 1.39e-10 |
| GO:0043226 | 1.79e-10 |
| GO:0044421 | 2.40e-09 |
| GO:0043231 | 8.57e-09 |
| GO:0005861 | 1.04e-08 |
| GO:0043227 | 1.63e-08 |
| GO:0044424 | 1.48e-07 |
| GO:0005634 | 7.50e-07 |
| GO:0003676 | 2.07e-06 |
| GO:0005201 | 7.90e-06 |
| GO:0097159 | 2.10e-05 |
| GO:1901363 | 2.84e-05 |
| GO:0004857 | 7.93e-05 |
| GO:0005518 | 9.43e-05 |
| GO:0005044 | 1.18e-04 |
| GO:0038024 | 1.64e-04 |
| GO:0008201 | 2.96e-04 |
| GO:0005244 | 3.86e-04 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06338
TATATTTATTAGTTCTAGAAAAACGTCTTCATATATATTTTTATTTGTAGATATGCTGACGGTCCTGATG
AAGATGTACAATCAGTCAATGAATATTCTATCGTTCTTCGTTTATTCCATGAAGTAAATGATTTGTAAAT
AGCAAATGATGCTCTTTATTATGCAGAAAAAAGCAATAATGTTTGTGCAGAGGCCTTGTTTATTTTTTGG
TTAGATACATTTTAGTAGATTAAAATGTATTATTTTCATATATGT
TATATTTATTAGTTCTAGAAAAACGTCTTCATATATATTTTTATTTGTAGATATGCTGACGGTCCTGATG
AAGATGTACAATCAGTCAATGAATATTCTATCGTTCTTCGTTTATTCCATGAAGTAAATGATTTGTAAAT
AGCAAATGATGCTCTTTATTATGCAGAAAAAAGCAATAATGTTTGTGCAGAGGCCTTGTTTATTTTTTGG
TTAGATACATTTTAGTAGATTAAAATGTATTATTTTCATATATGT