ZFLNCG06398
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562530 | liver | normal | 122.97 |
| SRR1035239 | liver | transgenic mCherry control | 81.81 |
| SRR700534 | heart | control morpholino | 24.82 |
| SRR700537 | heart | Gata4 morpholino | 15.21 |
| SRR516121 | skin | male and 3.5 year | 12.45 |
| SRR1562533 | testis | normal | 9.97 |
| SRR1035237 | liver | transgenic UHRF1 | 9.85 |
| SRR1562529 | intestine and pancreas | normal | 9.47 |
| SRR1647682 | spleen | normal | 9.16 |
| SRR516125 | skin | male and 3.5 year | 8.31 |
Express in tissues
Correlated coding gene
Download
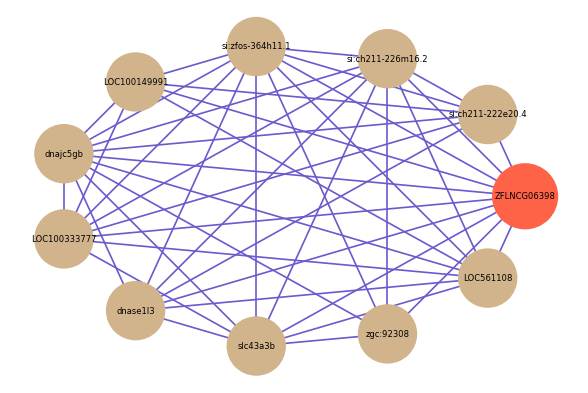
| gene | correlation coefficent |
|---|---|
| si:ch211-222e20.4 | 0.74 |
| slc43a3b | 0.61 |
| si:ch211-226m16.2 | 0.60 |
| si:zfos-364h11.1 | 0.60 |
| zgc:92308 | 0.59 |
| LOC100149991 | 0.58 |
| dnajc5gb | 0.58 |
| LOC100333777 | 0.58 |
| LOC561108 | 0.58 |
| dnase1l3 | 0.58 |
Gene Ontology
Download
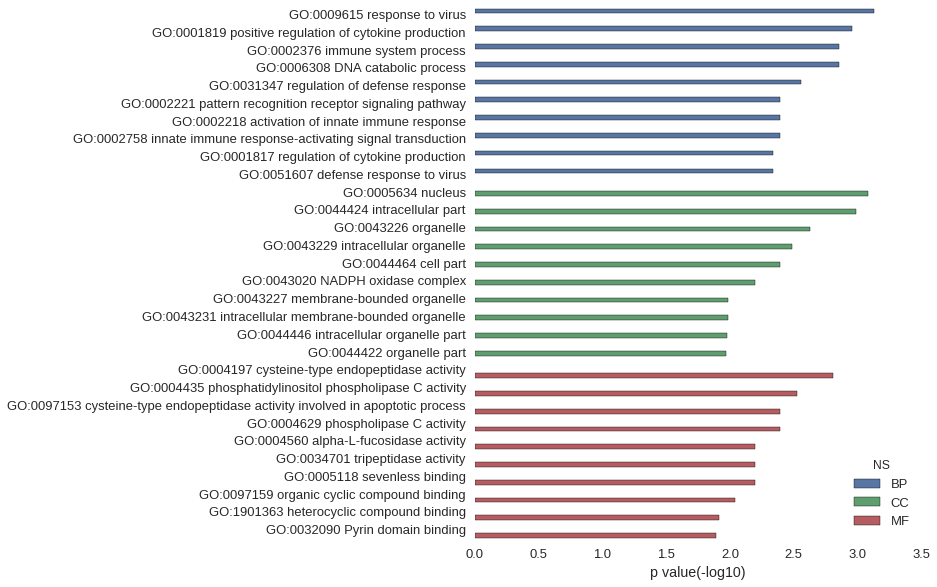
| GO | P value |
|---|---|
| GO:0009615 | 7.38e-04 |
| GO:0001819 | 1.10e-03 |
| GO:0002376 | 1.41e-03 |
| GO:0006308 | 1.41e-03 |
| GO:0031347 | 2.77e-03 |
| GO:0002221 | 4.02e-03 |
| GO:0002218 | 4.02e-03 |
| GO:0002758 | 4.02e-03 |
| GO:0001817 | 4.58e-03 |
| GO:0051607 | 4.58e-03 |
| GO:0005634 | 8.26e-04 |
| GO:0044424 | 1.03e-03 |
| GO:0043226 | 2.34e-03 |
| GO:0043229 | 3.27e-03 |
| GO:0044464 | 4.06e-03 |
| GO:0043020 | 6.40e-03 |
| GO:0043227 | 1.03e-02 |
| GO:0043231 | 1.04e-02 |
| GO:0044446 | 1.05e-02 |
| GO:0044422 | 1.07e-02 |
| GO:0004197 | 1.56e-03 |
| GO:0004435 | 3.01e-03 |
| GO:0097153 | 4.02e-03 |
| GO:0004629 | 4.02e-03 |
| GO:0004560 | 6.40e-03 |
| GO:0034701 | 6.40e-03 |
| GO:0005118 | 6.40e-03 |
| GO:0097159 | 9.06e-03 |
| GO:1901363 | 1.22e-02 |
| GO:0032090 | 1.28e-02 |
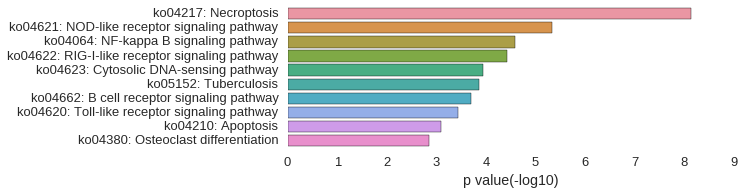
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06398
ACCCTGAGCACATCAACATGGTTTTGAAACGGATTACAGACTATCTTTTGTTTTGGACTTTTATTCATCT
TCCAAATACAGGTAATATACTTCATTTTTTTGAGTTTACTGTACTCTTTTAGTTAAAGCAAAGAAGCTGC
ATGTGATGTCATGATGTACATTACAAATAACAACTAATAATAAATAAAGGTCCCGGTGAGAGGCTTATTC
ATTTTATGTCTGGTTGGTTTTCTGTGATGTATTTTACAAAAAAAATGTGATGTATGTTTTGAATTGTCTG
AAGGGAGCATCCTGGTGTTTGTAACATTGAAATTTGTCAATCCACAGATGCAAAGAAAGTTATCAAAGCA
GTGGAAGGAGCCACTGTCCATCTGCCCTGCCATTTACCTCCTGTAGAGAGTCAACAAGTTACAGTAGAGT
GGACCATAAATACACACAGTAACAGTTCTATTTGTGCATGGATCACTGATAATAACTCTGTACACCCCTT
AAAAGATTGCTTTCCTCATTTTACATTAACCAGTAAGCCTATTAGATTAGAGATAAAAAATGTTCAGCCT
GGTGACTCTGGTAAATACAGCTGTAAAACAACAAGAGTAATTCCACCTCCGTCAGTGGAAGAAAGCTCAT
CTGTGATTCTCCGAGTCACAGATGATTTTGCTGTTCATTAGTGATTCTGAATTGTCTGTATGTTGAATAT
CATCTTACTTTGATGAATCTAATGTCCAGGTCTGTCCCTGAAGCCACTGAACAGCACTAATGACAGCTGT
GTTCATCTGCTCTGCTCTCTGGACTCTGATCTGGCCAACTTCACCTGGAGCCGAGGAGGTCAGATGTTAC
CACATGTGTCCACATTTAATGCTGATAATAGTGAACTGCAGCTGTGCAAACCTGACTGGAGTGAAGGAGA
CACAATCACCTGTCACGTCAACTACTCAGGAACCCAAACTCAAAAAAGCATCAAATTCAGCAGATCAGAA
AATGCTAACAGAGGTGAGTCAATTGAAAAACATATACTGAATCACTTTAGTAGCAAAATTTCATTATTAA
TCAGTTCACTGAAATGGTTCAATTGATTTTTAGATGTTATTTGTTTATTAATAAAATCTAAATTATTTAA
TTTAAAAAAAGTATAAATCAAACATTCCATGAGGGTAACCTATGGCCCATCTGTGGTCCGTCTTTCCCAG
GATACCAGGATTTTCCAGCTGTTTT
ACCCTGAGCACATCAACATGGTTTTGAAACGGATTACAGACTATCTTTTGTTTTGGACTTTTATTCATCT
TCCAAATACAGGTAATATACTTCATTTTTTTGAGTTTACTGTACTCTTTTAGTTAAAGCAAAGAAGCTGC
ATGTGATGTCATGATGTACATTACAAATAACAACTAATAATAAATAAAGGTCCCGGTGAGAGGCTTATTC
ATTTTATGTCTGGTTGGTTTTCTGTGATGTATTTTACAAAAAAAATGTGATGTATGTTTTGAATTGTCTG
AAGGGAGCATCCTGGTGTTTGTAACATTGAAATTTGTCAATCCACAGATGCAAAGAAAGTTATCAAAGCA
GTGGAAGGAGCCACTGTCCATCTGCCCTGCCATTTACCTCCTGTAGAGAGTCAACAAGTTACAGTAGAGT
GGACCATAAATACACACAGTAACAGTTCTATTTGTGCATGGATCACTGATAATAACTCTGTACACCCCTT
AAAAGATTGCTTTCCTCATTTTACATTAACCAGTAAGCCTATTAGATTAGAGATAAAAAATGTTCAGCCT
GGTGACTCTGGTAAATACAGCTGTAAAACAACAAGAGTAATTCCACCTCCGTCAGTGGAAGAAAGCTCAT
CTGTGATTCTCCGAGTCACAGATGATTTTGCTGTTCATTAGTGATTCTGAATTGTCTGTATGTTGAATAT
CATCTTACTTTGATGAATCTAATGTCCAGGTCTGTCCCTGAAGCCACTGAACAGCACTAATGACAGCTGT
GTTCATCTGCTCTGCTCTCTGGACTCTGATCTGGCCAACTTCACCTGGAGCCGAGGAGGTCAGATGTTAC
CACATGTGTCCACATTTAATGCTGATAATAGTGAACTGCAGCTGTGCAAACCTGACTGGAGTGAAGGAGA
CACAATCACCTGTCACGTCAACTACTCAGGAACCCAAACTCAAAAAAGCATCAAATTCAGCAGATCAGAA
AATGCTAACAGAGGTGAGTCAATTGAAAAACATATACTGAATCACTTTAGTAGCAAAATTTCATTATTAA
TCAGTTCACTGAAATGGTTCAATTGATTTTTAGATGTTATTTGTTTATTAATAAAATCTAAATTATTTAA
TTTAAAAAAAGTATAAATCAAACATTCCATGAGGGTAACCTATGGCCCATCTGTGGTCCGTCTTTCCCAG
GATACCAGGATTTTCCAGCTGTTTT