ZFLNCG06405
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR700534 | heart | control morpholino | 51.33 |
| SRR700537 | heart | Gata4 morpholino | 34.65 |
| SRR1647684 | spleen | SVCV treatment | 25.91 |
| ERR023143 | swim bladder | normal | 12.28 |
| SRR1647680 | head kidney | SVCV treatment | 11.64 |
| SRR594769 | blood | normal | 11.60 |
| SRR957180 | heart | 7 days after heart tip amputation | 11.05 |
| SRR1647679 | head kidney | normal | 10.89 |
| SRR1647683 | spleen | SVCV treatment | 9.96 |
| SRR1647681 | head kidney | SVCV treatment | 9.52 |
Express in tissues
Correlated coding gene
Download
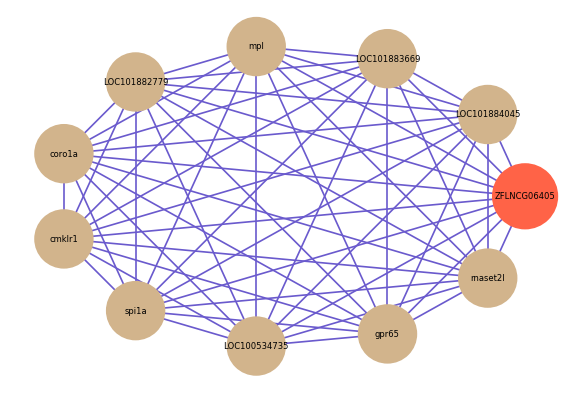
| gene | correlation coefficent |
|---|---|
| LOC101884045 | 0.60 |
| LOC101883669 | 0.56 |
| mpl | 0.56 |
| LOC101882779 | 0.56 |
| coro1a | 0.56 |
| cmklr1 | 0.55 |
| spi1a | 0.55 |
| LOC100534735 | 0.55 |
| gpr65 | 0.55 |
| rnaset2l | 0.55 |
Gene Ontology
Download
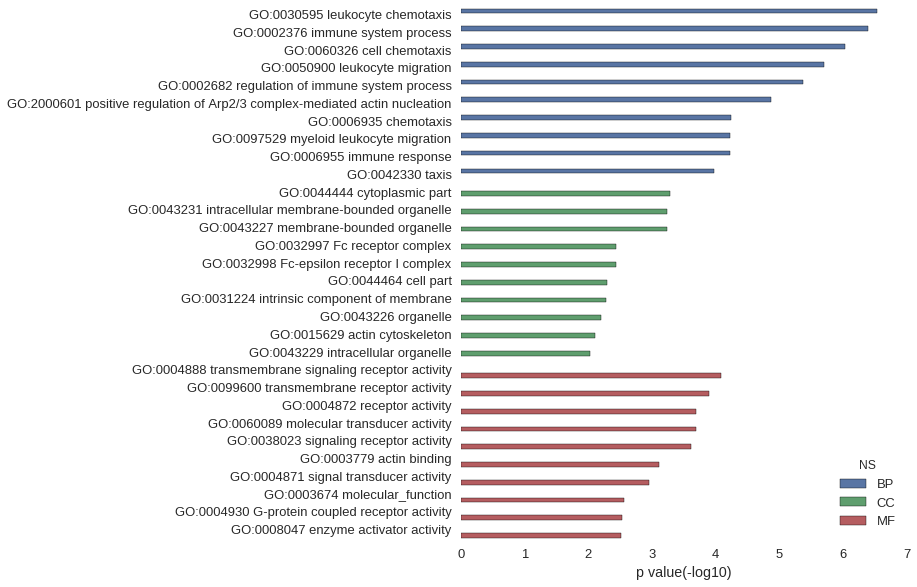
| GO | P value |
|---|---|
| GO:0030595 | 2.92e-07 |
| GO:0002376 | 4.15e-07 |
| GO:0060326 | 9.47e-07 |
| GO:0050900 | 1.98e-06 |
| GO:0002682 | 4.33e-06 |
| GO:2000601 | 1.34e-05 |
| GO:0006935 | 5.67e-05 |
| GO:0097529 | 6.04e-05 |
| GO:0006955 | 6.04e-05 |
| GO:0042330 | 1.07e-04 |
| GO:0044444 | 5.20e-04 |
| GO:0043231 | 5.74e-04 |
| GO:0043227 | 5.79e-04 |
| GO:0032997 | 3.70e-03 |
| GO:0032998 | 3.70e-03 |
| GO:0044464 | 4.99e-03 |
| GO:0031224 | 5.29e-03 |
| GO:0043226 | 6.27e-03 |
| GO:0015629 | 7.79e-03 |
| GO:0043229 | 9.41e-03 |
| GO:0004888 | 8.22e-05 |
| GO:0099600 | 1.29e-04 |
| GO:0004872 | 2.03e-04 |
| GO:0060089 | 2.03e-04 |
| GO:0038023 | 2.47e-04 |
| GO:0003779 | 7.79e-04 |
| GO:0004871 | 1.10e-03 |
| GO:0003674 | 2.70e-03 |
| GO:0004930 | 2.99e-03 |
| GO:0008047 | 3.05e-03 |
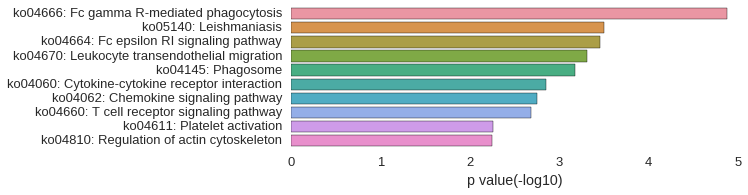
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06405
GTTAATGCGGTAAAGTTGCAATGTTTTTGCTGTCATCTAGCGGTTGATCATAATGTAACATTTACTTCCC
CCATAGATTACGCTTGGTGTTTTGACAAAAGATTCATAATAATTTTGAAAGGGTGTTTCGCTTTTTCACT
CCAGCGTTTGTTGTAGTTGTCATTTCAGTTATTCCTCTTCTCTTGTCTTGTGAGCTGTCGAGAGACACAA
TCCTAAACGCCGGTCTTCTCCATGACCGTGGCTCCAACCTCAGAGTAGCCGAAGTCGTCGAGTTGTTGCT
GTGCAGCGGAGGCCTGACTGTTCTCTTTCGGCTTAATGAAGTCTGTGTAGACGCACGGAAGAGGAATCAC
TTTTTCGTTGTCTG
GTTAATGCGGTAAAGTTGCAATGTTTTTGCTGTCATCTAGCGGTTGATCATAATGTAACATTTACTTCCC
CCATAGATTACGCTTGGTGTTTTGACAAAAGATTCATAATAATTTTGAAAGGGTGTTTCGCTTTTTCACT
CCAGCGTTTGTTGTAGTTGTCATTTCAGTTATTCCTCTTCTCTTGTCTTGTGAGCTGTCGAGAGACACAA
TCCTAAACGCCGGTCTTCTCCATGACCGTGGCTCCAACCTCAGAGTAGCCGAAGTCGTCGAGTTGTTGCT
GTGCAGCGGAGGCCTGACTGTTCTCTTTCGGCTTAATGAAGTCTGTGTAGACGCACGGAAGAGGAATCAC
TTTTTCGTTGTCTG