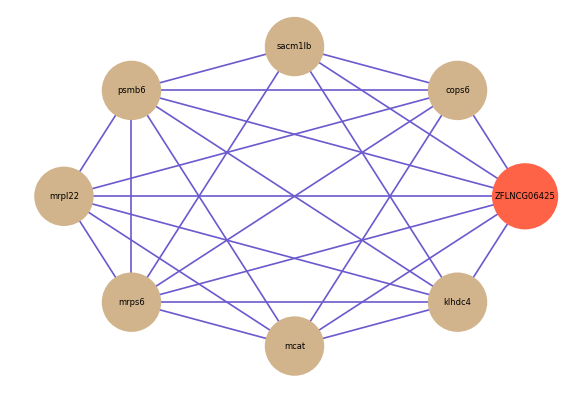
ZFLNCG06425
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR748488 | sperm | normal | 276.99 |
| SRR1005530 | embryo | RPL11 morpholino | 86.54 |
| SRR801555 | embryo | RPS19 morpholino | 78.61 |
| SRR1035978 | 13 hpf | rx3-/- | 64.96 |
| SRR801556 | embryo | RPS19 and p53 morpholino | 57.77 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 47.99 |
| SRR658543 | 6 somite | Gata5 morphan | 33.35 |
| SRR1004787 | larvae | impdh1a morpholino | 29.35 |
| SRR1035237 | liver | transgenic UHRF1 | 29.22 |
| SRR658541 | 6 somite | normal | 27.91 |
Express in tissues
Correlated coding gene
Download
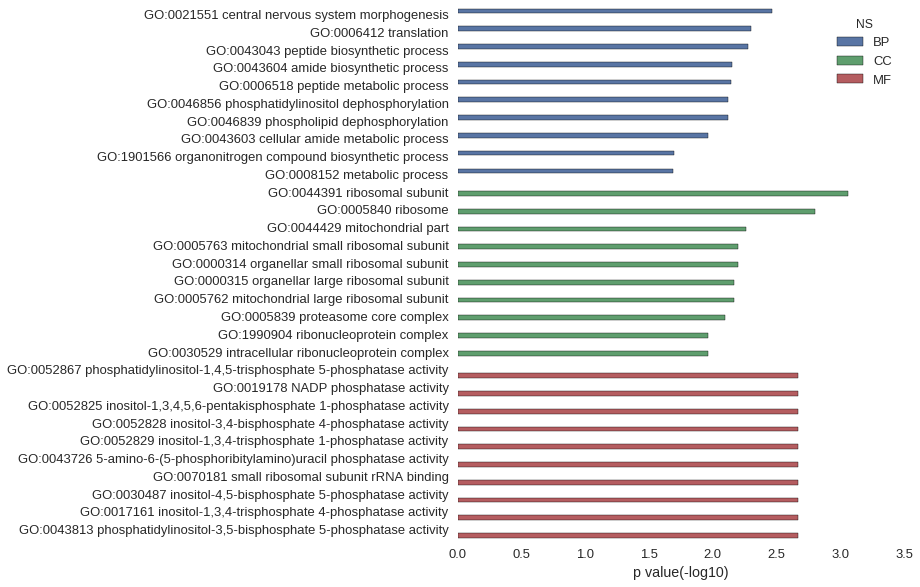
Gene Ontology
Download
| GO | P value |
|---|---|
| GO:0021551 | 3.41e-03 |
| GO:0006412 | 5.04e-03 |
| GO:0043043 | 5.27e-03 |
| GO:0043604 | 7.10e-03 |
| GO:0006518 | 7.23e-03 |
| GO:0046856 | 7.65e-03 |
| GO:0046839 | 7.65e-03 |
| GO:0043603 | 1.08e-02 |
| GO:1901566 | 2.03e-02 |
| GO:0008152 | 2.06e-02 |
| GO:0044391 | 8.74e-04 |
| GO:0005840 | 1.58e-03 |
| GO:0044429 | 5.46e-03 |
| GO:0005763 | 6.38e-03 |
| GO:0000314 | 6.38e-03 |
| GO:0000315 | 6.80e-03 |
| GO:0005762 | 6.80e-03 |
| GO:0005839 | 8.08e-03 |
| GO:1990904 | 1.08e-02 |
| GO:0030529 | 1.08e-02 |
| GO:0052867 | 2.13e-03 |
| GO:0019178 | 2.13e-03 |
| GO:0052825 | 2.13e-03 |
| GO:0052828 | 2.13e-03 |
| GO:0052829 | 2.13e-03 |
| GO:0043726 | 2.13e-03 |
| GO:0070181 | 2.13e-03 |
| GO:0030487 | 2.13e-03 |
| GO:0017161 | 2.13e-03 |
| GO:0043813 | 2.13e-03 |
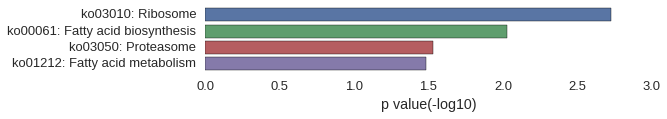
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06425
GACTATAGCATATACGATATAAGACTTGCTGTATAGCAGAAGATCTGCGTTTCTGTAACTCGAGTCGCAC
TTAGTTCACTTCACTCCAGTGACCCGCTGGCAGCTTTACTTCTGGCTGTCCCGGTGCATCAAACTTCCCA
GACTGACTTGACAATCCCCTGTATATGTTTAAAAATATGCAGCACCTATTAAATGAGATGTCTGTATGAA
ATGAATCCTCATAAAGAGGTTGTTGTCTGAAAGAATGAATTAACTTACTAGGGATCGAGTGCCAGATGGC
TATTCAGCACTGCGCATGCGTACAAG
GACTATAGCATATACGATATAAGACTTGCTGTATAGCAGAAGATCTGCGTTTCTGTAACTCGAGTCGCAC
TTAGTTCACTTCACTCCAGTGACCCGCTGGCAGCTTTACTTCTGGCTGTCCCGGTGCATCAAACTTCCCA
GACTGACTTGACAATCCCCTGTATATGTTTAAAAATATGCAGCACCTATTAAATGAGATGTCTGTATGAA
ATGAATCCTCATAAAGAGGTTGTTGTCTGAAAGAATGAATTAACTTACTAGGGATCGAGTGCCAGATGGC
TATTCAGCACTGCGCATGCGTACAAG