ZFLNCG06492
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1648854 | brain | normal | 61.39 |
| SRR1648856 | brain | normal | 46.89 |
| ERR023144 | brain | normal | 23.66 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 18.11 |
| SRR516135 | skin | male and 5 month | 15.59 |
| SRR1648855 | brain | normal | 14.93 |
| SRR1523211 | embryo | control morpholino | 13.36 |
| SRR038625 | embryo | traf6 morpholino | 13.33 |
| SRR1291414 | 5 dpi | normal | 13.12 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 12.59 |
Express in tissues
Correlated coding gene
Download
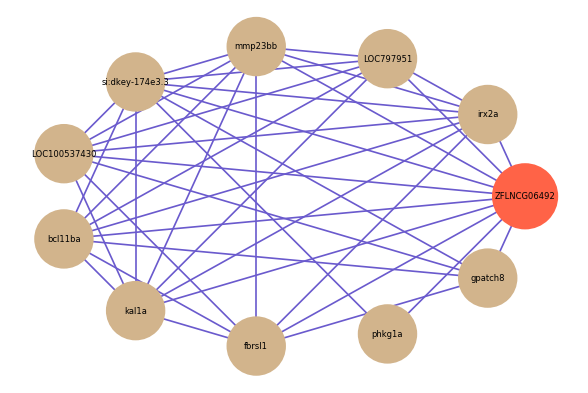
| gene | correlation coefficent |
|---|---|
| irx2a | 0.56 |
| LOC797951 | 0.55 |
| mmp23bb | 0.54 |
| si:dkey-174e3.3 | 0.54 |
| LOC100537430 | 0.53 |
| bcl11ba | 0.52 |
| phkg1a | 0.51 |
| kal1a | 0.51 |
| fbrsl1 | 0.50 |
| gpatch8 | 0.50 |
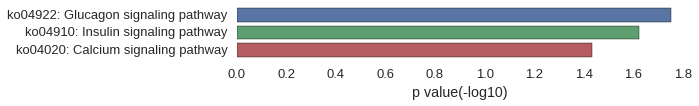
Gene Ontology
Download
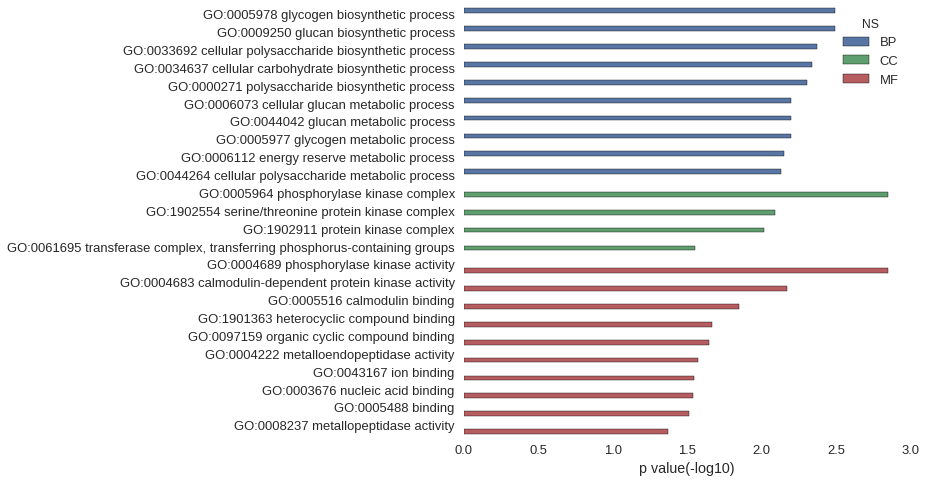
| GO | P value |
|---|---|
| GO:0005978 | 3.19e-03 |
| GO:0009250 | 3.19e-03 |
| GO:0033692 | 4.26e-03 |
| GO:0034637 | 4.61e-03 |
| GO:0000271 | 4.97e-03 |
| GO:0006073 | 6.38e-03 |
| GO:0044042 | 6.38e-03 |
| GO:0005977 | 6.38e-03 |
| GO:0006112 | 7.09e-03 |
| GO:0044264 | 7.44e-03 |
| GO:0005964 | 1.42e-03 |
| GO:1902554 | 8.15e-03 |
| GO:1902911 | 9.56e-03 |
| GO:0061695 | 2.78e-02 |
| GO:0004689 | 1.42e-03 |
| GO:0004683 | 6.73e-03 |
| GO:0005516 | 1.41e-02 |
| GO:1901363 | 2.16e-02 |
| GO:0097159 | 2.24e-02 |
| GO:0004222 | 2.67e-02 |
| GO:0043167 | 2.83e-02 |
| GO:0003676 | 2.89e-02 |
| GO:0005488 | 3.07e-02 |
| GO:0008237 | 4.23e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06492
GTGTGCTACGGTACGGTACGAGTCGGTACTTTCAATAGGTACCAAAAAGCAAGTTACCATAGGTACTAAA
CCGTTACTGTTACTCCTGCTGTTCTATTGACGCTCCCCTGCTGGATTACCTCTTTTATTCATTCATTCAT
TTTCCTTCGACTTAGTGTTTATTTCAGAGGTCACCACTGCGGAATGAACTGCCAACTATTTCAGCATATG
TTTTATGCTGCAGATGCCTTTCCAACTGAAT
GTGTGCTACGGTACGGTACGAGTCGGTACTTTCAATAGGTACCAAAAAGCAAGTTACCATAGGTACTAAA
CCGTTACTGTTACTCCTGCTGTTCTATTGACGCTCCCCTGCTGGATTACCTCTTTTATTCATTCATTCAT
TTTCCTTCGACTTAGTGTTTATTTCAGAGGTCACCACTGCGGAATGAACTGCCAACTATTTCAGCATATG
TTTTATGCTGCAGATGCCTTTCCAACTGAAT