ZFLNCG06520
Basic Information
Chromesome: chr10
Start: 41973806
End: 41974822
Transcript: ZFLNCT09987
Known as: ENSDARG00000095264
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 0.99 |
| SRR1562533 | testis | normal | 0.42 |
| SRR1342217 | embryo | infection with Mycobacterium marinum | 0.39 |
| SRR1028002 | head | normal | 0.31 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 0.31 |
| SRR1205154 | 5dpf | transgenic sqET20 | 0.28 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 0.25 |
| SRR1028003 | head | normal | 0.25 |
| SRR1167753 | embryo | bacterial infection | 0.24 |
| SRR1523211 | embryo | control morpholino | 0.23 |
Express in tissues
Correlated coding gene
Download
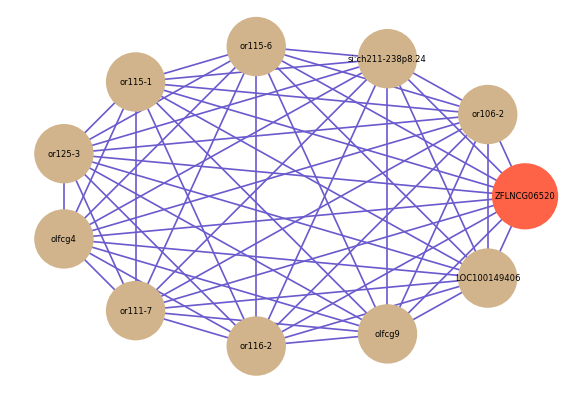
| gene | correlation coefficent |
|---|---|
| or106-2 | 0.62 |
| si:ch211-238p8.24 | 0.62 |
| or115-6 | 0.62 |
| or115-1 | 0.61 |
| or125-3 | 0.61 |
| olfcg4 | 0.60 |
| or111-7 | 0.59 |
| or116-2 | 0.59 |
| olfcg9 | 0.58 |
| LOC100149406 | 0.58 |
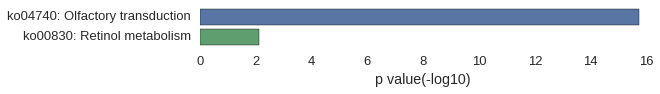
Gene Ontology
Download
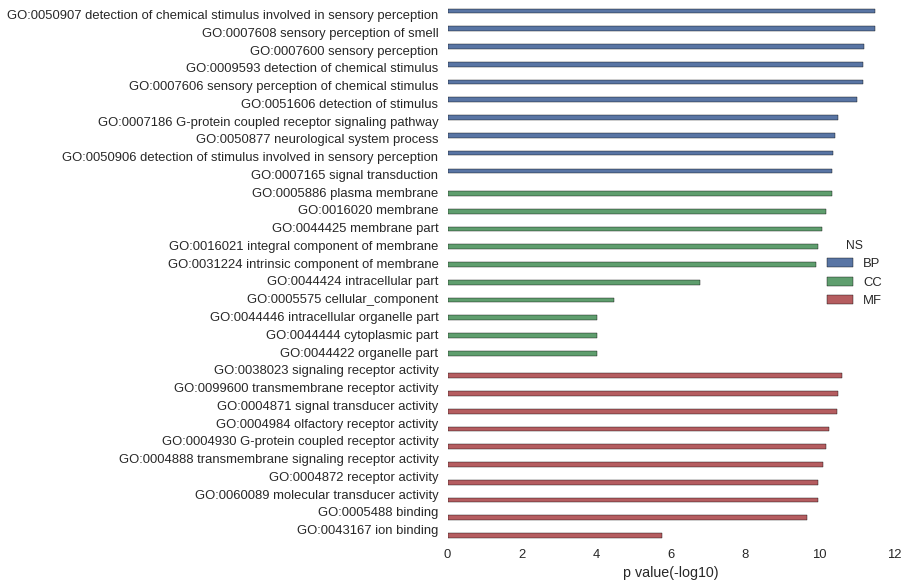
| GO | P value |
|---|---|
| GO:0050907 | 3.24e-12 |
| GO:0007608 | 3.24e-12 |
| GO:0007600 | 6.39e-12 |
| GO:0009593 | 6.62e-12 |
| GO:0007606 | 6.62e-12 |
| GO:0051606 | 9.58e-12 |
| GO:0007186 | 3.25e-11 |
| GO:0050877 | 3.85e-11 |
| GO:0050906 | 4.28e-11 |
| GO:0007165 | 4.72e-11 |
| GO:0005886 | 4.61e-11 |
| GO:0016020 | 6.68e-11 |
| GO:0044425 | 8.73e-11 |
| GO:0016021 | 1.08e-10 |
| GO:0031224 | 1.24e-10 |
| GO:0044424 | 1.64e-07 |
| GO:0005575 | 3.34e-05 |
| GO:0044446 | 9.33e-05 |
| GO:0044444 | 9.34e-05 |
| GO:0044422 | 9.69e-05 |
| GO:0038023 | 2.42e-11 |
| GO:0099600 | 3.26e-11 |
| GO:0004871 | 3.43e-11 |
| GO:0004984 | 5.65e-11 |
| GO:0004930 | 6.57e-11 |
| GO:0004888 | 7.85e-11 |
| GO:0004872 | 1.08e-10 |
| GO:0060089 | 1.08e-10 |
| GO:0005488 | 2.11e-10 |
| GO:0043167 | 1.69e-06 |
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT09987 | ENST00000421055; ENST00000486251; NONHSAT024249; NONHSAT152762; NONHSAT104458; NONHSAT125668; NONHSAT125667; NONHSAT008869; | ENSMUST00000134011; ENSMUST00000172211; ENSMUST00000120418; NONMMUT124900; NONMMUT085607; NONMMUT060621; | Collinearity with conserved coding gene |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06520
ACAAACTCCTAACAGGGGGAGACTCACTCATGGCCTATGAGACAGAGGATCAGGAGACTCAATACTGCTT
TCCTAACATCAACTCATCATGTGTCAAGGAGAATCGATCCAGTTATGAATATTATATCATGTATGTGTTT
TTTTCATTGCTGTCAGCATGGACTGTTTCTGAACCTGCTGGTGATCATCTCCATCTCTCACTACAAGAAT
CTTCACACTCCAACCAACATGATTATTCTCTCTCTGGCTGTTGCTGATATGCTAATGGGACTTATTGTGA
TGCCTGTGGATGCCATTAAGCTGATTGAGACCTGTTGGTACTTTGGAGAAAGTTTGTGTGATGTGTTTAT
AATAATTATGGGTCTACTTCTCTCAACATCTCTTGGTAATTTAGTTTTAATTGCTGTTGATCGTTATGTA
GCTGTGTGTCACCCTTTAGTGTATCCACTAAAAATAACAATGACTAAAATGTTAATCATTGTCAGTTGCT
GCTGGCTTTTTTCTTCAGCTTATAATATTACAATTGTAAAAAGTACCTCACAGAGAAAAGATGGGTGTTA
CAGCGAATGTAATGCTACCCTTACATATGAATGGAGAGCTATTGACTTAATATTTTCATTCCTGCTTCCT
TGTACAATTATCATAACTTTACATTTGCGAATATTTTATGTTGTACATCAGCAAGTGAAAGTGATCAACT
CTCAAATGAAGCGAGGAAAATCTGTCATGGAAAGTTCAGTGAAGAGAAAATCTGAGAGCAAAGCAGCTCT
GACATTAGGAATAATTGTAGCTATTCATCTGCTTTGCTGGATCCCATACTATATATTGACTCTAACTGAT
AACATAGCTATTCCACCCACCGTATTATATTGTCTAATATGGGTCTTATACATTAATTCATGTCTGAATC
CTATTCTCTATGCTTTCTTTTACCCCTGGTTTAGAAAATCATTCAAACACATCTTAACTCTTAAAATATT
TCAGCTAGCATCCTCTCATGTCAATCTTTTCACAAG
ACAAACTCCTAACAGGGGGAGACTCACTCATGGCCTATGAGACAGAGGATCAGGAGACTCAATACTGCTT
TCCTAACATCAACTCATCATGTGTCAAGGAGAATCGATCCAGTTATGAATATTATATCATGTATGTGTTT
TTTTCATTGCTGTCAGCATGGACTGTTTCTGAACCTGCTGGTGATCATCTCCATCTCTCACTACAAGAAT
CTTCACACTCCAACCAACATGATTATTCTCTCTCTGGCTGTTGCTGATATGCTAATGGGACTTATTGTGA
TGCCTGTGGATGCCATTAAGCTGATTGAGACCTGTTGGTACTTTGGAGAAAGTTTGTGTGATGTGTTTAT
AATAATTATGGGTCTACTTCTCTCAACATCTCTTGGTAATTTAGTTTTAATTGCTGTTGATCGTTATGTA
GCTGTGTGTCACCCTTTAGTGTATCCACTAAAAATAACAATGACTAAAATGTTAATCATTGTCAGTTGCT
GCTGGCTTTTTTCTTCAGCTTATAATATTACAATTGTAAAAAGTACCTCACAGAGAAAAGATGGGTGTTA
CAGCGAATGTAATGCTACCCTTACATATGAATGGAGAGCTATTGACTTAATATTTTCATTCCTGCTTCCT
TGTACAATTATCATAACTTTACATTTGCGAATATTTTATGTTGTACATCAGCAAGTGAAAGTGATCAACT
CTCAAATGAAGCGAGGAAAATCTGTCATGGAAAGTTCAGTGAAGAGAAAATCTGAGAGCAAAGCAGCTCT
GACATTAGGAATAATTGTAGCTATTCATCTGCTTTGCTGGATCCCATACTATATATTGACTCTAACTGAT
AACATAGCTATTCCACCCACCGTATTATATTGTCTAATATGGGTCTTATACATTAATTCATGTCTGAATC
CTATTCTCTATGCTTTCTTTTACCCCTGGTTTAGAAAATCATTCAAACACATCTTAACTCTTAAAATATT
TCAGCTAGCATCCTCTCATGTCAATCTTTTCACAAG