ZFLNCG06526
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592701 | pineal gland | normal | 102.32 |
| SRR592703 | pineal gland | normal | 82.42 |
| SRR592702 | pineal gland | normal | 64.80 |
| SRR592700 | pineal gland | normal | 30.22 |
| SRR594771 | endothelium | normal | 22.50 |
| ERR023143 | swim bladder | normal | 22.47 |
| SRR592699 | pineal gland | normal | 22.17 |
| SRR1048059 | pineal gland | light | 20.55 |
| SRR1035239 | liver | transgenic mCherry control | 18.57 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 18.30 |
Express in tissues
Correlated coding gene
Download
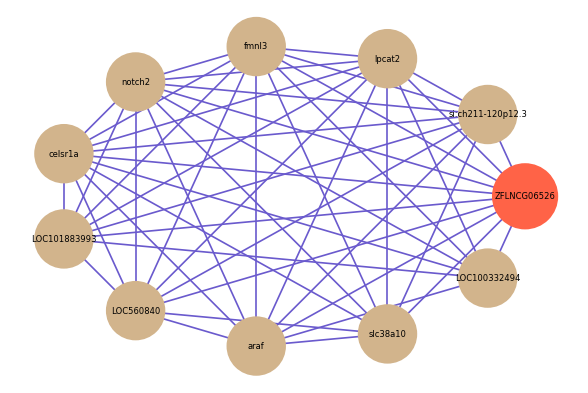
| gene | correlation coefficent |
|---|---|
| si:ch211-120p12.3 | 0.64 |
| lpcat2 | 0.61 |
| fmnl3 | 0.61 |
| notch2 | 0.60 |
| celsr1a | 0.60 |
| LOC101883993 | 0.60 |
| LOC560840 | 0.60 |
| araf | 0.59 |
| LOC100332494 | 0.59 |
| slc38a10 | 0.59 |
Gene Ontology
Download
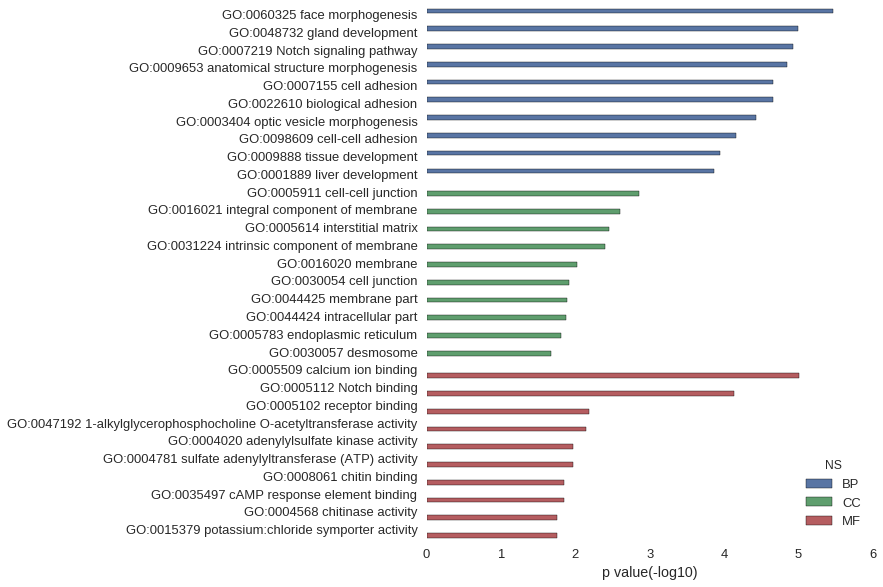
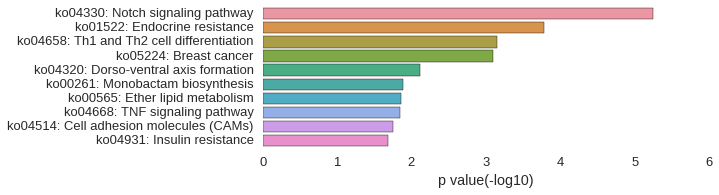
| GO | P value |
|---|---|
| GO:0060325 | 3.49e-06 |
| GO:0048732 | 1.01e-05 |
| GO:0007219 | 1.17e-05 |
| GO:0009653 | 1.43e-05 |
| GO:0007155 | 2.22e-05 |
| GO:0022610 | 2.22e-05 |
| GO:0003404 | 3.70e-05 |
| GO:0098609 | 6.83e-05 |
| GO:0009888 | 1.13e-04 |
| GO:0001889 | 1.35e-04 |
| GO:0005911 | 1.40e-03 |
| GO:0016021 | 2.50e-03 |
| GO:0005614 | 3.55e-03 |
| GO:0031224 | 3.94e-03 |
| GO:0016020 | 9.47e-03 |
| GO:0030054 | 1.23e-02 |
| GO:0044425 | 1.30e-02 |
| GO:0044424 | 1.34e-02 |
| GO:0005783 | 1.57e-02 |
| GO:0030057 | 2.11e-02 |
| GO:0005509 | 9.79e-06 |
| GO:0005112 | 7.39e-05 |
| GO:0005102 | 6.54e-03 |
| GO:0047192 | 7.09e-03 |
| GO:0004020 | 1.06e-02 |
| GO:0004781 | 1.06e-02 |
| GO:0008061 | 1.41e-02 |
| GO:0035497 | 1.41e-02 |
| GO:0004568 | 1.76e-02 |
| GO:0015379 | 1.76e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06526
TGATATAAAAAGGCTTGTATAAACCATTACAAAGAGCATCAAATAACGAAAAGTATGAATACACGGAGAT
TATAAGCCTGTATATGATAACAAAAAGATGAAAGCGCTCTACACATCATGGTAGAAGATGGTAGTGAAGC
TGAGCTAAAGTGCAGCAGAGACTCCAAACGAGGCGTCAGAAAGACTGAGGTAGCCGATACATGATGACTA
CACCAGACCTGCTTCCACCACAGGACGACTAAACAGGGCGTTCATAACAGAGATAGATCACCCAAAAACG
AAAATTCT
TGATATAAAAAGGCTTGTATAAACCATTACAAAGAGCATCAAATAACGAAAAGTATGAATACACGGAGAT
TATAAGCCTGTATATGATAACAAAAAGATGAAAGCGCTCTACACATCATGGTAGAAGATGGTAGTGAAGC
TGAGCTAAAGTGCAGCAGAGACTCCAAACGAGGCGTCAGAAAGACTGAGGTAGCCGATACATGATGACTA
CACCAGACCTGCTTCCACCACAGGACGACTAAACAGGGCGTTCATAACAGAGATAGATCACCCAAAAACG
AAAATTCT