ZFLNCG06561
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR891511 | brain | normal | 152.71 |
| SRR1035237 | liver | transgenic UHRF1 | 142.48 |
| SRR658539 | bud | Gata5/6 morphant | 133.87 |
| SRR372802 | 5 dpf | normal | 125.75 |
| SRR372796 | bud | normal | 125.67 |
| SRR801555 | embryo | RPS19 morpholino | 119.49 |
| SRR592700 | pineal gland | normal | 119.27 |
| SRR658543 | 6 somite | Gata5 morphan | 117.78 |
| SRR658535 | bud | Gata5 morphant | 116.72 |
| SRR519752 | 7 dpf | VD3 treatment | 114.62 |
Express in tissues
Correlated coding gene
Download
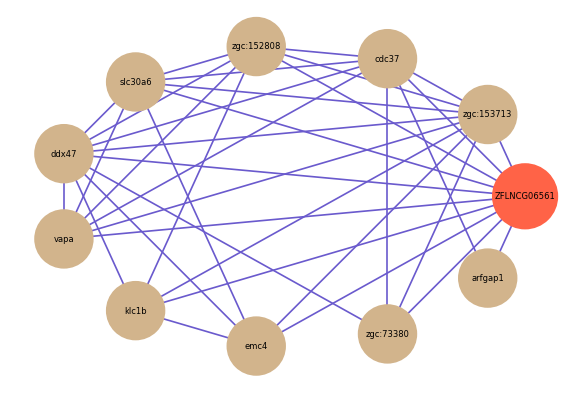
| gene | correlation coefficent |
|---|---|
| zgc:153713 | 0.70 |
| cdc37 | 0.67 |
| arfgap1 | 0.65 |
| zgc:152808 | 0.65 |
| slc30a6 | 0.64 |
| ddx47 | 0.64 |
| vapa | 0.62 |
| klc1b | 0.60 |
| emc4 | 0.59 |
| zgc:73380 | 0.59 |
Gene Ontology
Download
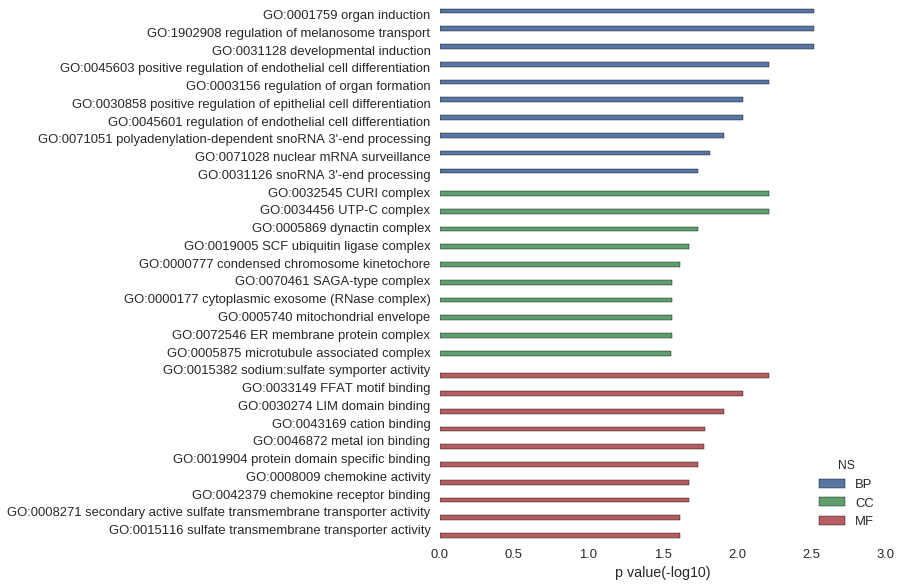
| GO | P value |
|---|---|
| GO:0001759 | 3.06e-03 |
| GO:1902908 | 3.06e-03 |
| GO:0031128 | 3.06e-03 |
| GO:0045603 | 6.10e-03 |
| GO:0003156 | 6.10e-03 |
| GO:0030858 | 9.14e-03 |
| GO:0045601 | 9.14e-03 |
| GO:0071051 | 1.22e-02 |
| GO:0071028 | 1.52e-02 |
| GO:0031126 | 1.82e-02 |
| GO:0032545 | 6.10e-03 |
| GO:0034456 | 6.10e-03 |
| GO:0005869 | 1.82e-02 |
| GO:0019005 | 2.12e-02 |
| GO:0000777 | 2.42e-02 |
| GO:0070461 | 2.72e-02 |
| GO:0000177 | 2.72e-02 |
| GO:0005740 | 2.72e-02 |
| GO:0072546 | 2.72e-02 |
| GO:0005875 | 2.77e-02 |
| GO:0015382 | 6.10e-03 |
| GO:0033149 | 9.14e-03 |
| GO:0030274 | 1.22e-02 |
| GO:0043169 | 1.65e-02 |
| GO:0046872 | 1.68e-02 |
| GO:0019904 | 1.83e-02 |
| GO:0008009 | 2.12e-02 |
| GO:0042379 | 2.12e-02 |
| GO:0008271 | 2.42e-02 |
| GO:0015116 | 2.42e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06561
CAGAATGCCGCGGTCTGCCAGCGCAGCAGAGATCAATGGTCGGCTGCTGATCAGAGGAGCCGCTGATTAT
TGAGATTAAAGGCGTTAATCAGCGCAGAGCCTCCGGTGCCGAAGCGCTGACAGCGGCACAGAGAGCAGGA
GCAGCCGCCGCCATCACCATCATCATCACCTTCATCACCACAGCTGAACAACAGCCCAGCCGGTGAACGA
CAGTGAGTATACGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTCGCGCTTCATGATTTGCCGGCGAACATC
CAATAATCACCTGTAACGACAGCGACACACACACACACACACACACATACACACATACACATTGAAAATC
TTCATTGCTCAGATTCCACACATACACACACACCTCACAGACATGTTTTCACTCACACACACACACACAC
ACACACACACACACACTGTTATGATTGTTTTCTTGATCGATGGTGTTTATTGATTGATTATAGAAGAGTG
TCAGATATGGGGGAATTATTGTTCATCAGTAGTCAATAACACACACACACACACACACACACACACACAT
TAAATGACGACACTACACGTGATTCAGTGTGAGATGTCAGTATAGGGTCAGGAGATGTTTGAGATGACGT
CATGAGTGTTATGAATGTGATCAGTCAGTATTCGGGTCTGTAATATGTCAGACAGTCATTATTGTTGTGA
TTATGGATGAAGAGTTCTTCACACACTGCTGTAATATTTCATAAATCGTGTATTAATCTCTGTCATGTTG
ATGTGGGACTCTCCTGTGTGTGTGTTTAGAAGATAACCCATGTTGAATGAGTTGTGTCAGTGTTGATATC
CTGCAGCTCTGAGGAGGAGGCGCTTCTTCATCATCATGTTCATTAGAGCGCTGCTGCATGATGATGATGA
TGATGATGATGATGATGATGATGATGATGAGCTGTTCACTCATACAGTAATGCACAGCGGCTCATCCTGA
TCTACAGCAACACTCCACCGACCCGCAGCTGAC
CAGAATGCCGCGGTCTGCCAGCGCAGCAGAGATCAATGGTCGGCTGCTGATCAGAGGAGCCGCTGATTAT
TGAGATTAAAGGCGTTAATCAGCGCAGAGCCTCCGGTGCCGAAGCGCTGACAGCGGCACAGAGAGCAGGA
GCAGCCGCCGCCATCACCATCATCATCACCTTCATCACCACAGCTGAACAACAGCCCAGCCGGTGAACGA
CAGTGAGTATACGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTCGCGCTTCATGATTTGCCGGCGAACATC
CAATAATCACCTGTAACGACAGCGACACACACACACACACACACACATACACACATACACATTGAAAATC
TTCATTGCTCAGATTCCACACATACACACACACCTCACAGACATGTTTTCACTCACACACACACACACAC
ACACACACACACACACTGTTATGATTGTTTTCTTGATCGATGGTGTTTATTGATTGATTATAGAAGAGTG
TCAGATATGGGGGAATTATTGTTCATCAGTAGTCAATAACACACACACACACACACACACACACACACAT
TAAATGACGACACTACACGTGATTCAGTGTGAGATGTCAGTATAGGGTCAGGAGATGTTTGAGATGACGT
CATGAGTGTTATGAATGTGATCAGTCAGTATTCGGGTCTGTAATATGTCAGACAGTCATTATTGTTGTGA
TTATGGATGAAGAGTTCTTCACACACTGCTGTAATATTTCATAAATCGTGTATTAATCTCTGTCATGTTG
ATGTGGGACTCTCCTGTGTGTGTGTTTAGAAGATAACCCATGTTGAATGAGTTGTGTCAGTGTTGATATC
CTGCAGCTCTGAGGAGGAGGCGCTTCTTCATCATCATGTTCATTAGAGCGCTGCTGCATGATGATGATGA
TGATGATGATGATGATGATGATGATGATGAGCTGTTCACTCATACAGTAATGCACAGCGGCTCATCCTGA
TCTACAGCAACACTCCACCGACCCGCAGCTGAC
