ZFLNCG06644
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 46.97 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 34.69 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 34.27 |
| SRR535978 | larvae | normal | 31.48 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 28.82 |
| ERR023143 | swim bladder | normal | 26.20 |
| SRR700537 | heart | Gata4 morpholino | 24.96 |
| SRR516125 | skin | male and 3.5 year | 24.64 |
| SRR516128 | skin | male and 5 month | 24.34 |
| SRR700534 | heart | control morpholino | 23.49 |
Express in tissues
Correlated coding gene
Download
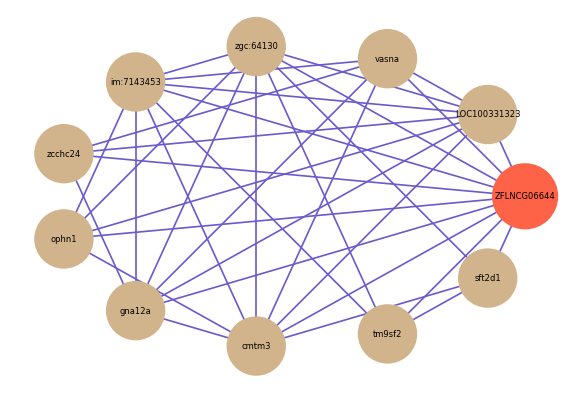
| gene | correlation coefficent |
|---|---|
| LOC100331323 | 0.61 |
| vasna | 0.59 |
| zgc:64130 | 0.59 |
| im:7143453 | 0.58 |
| tm9sf2 | 0.57 |
| sft2d1 | 0.56 |
| zcchc24 | 0.56 |
| ophn1 | 0.55 |
| gna12a | 0.55 |
| cmtm3 | 0.55 |
Gene Ontology
Download
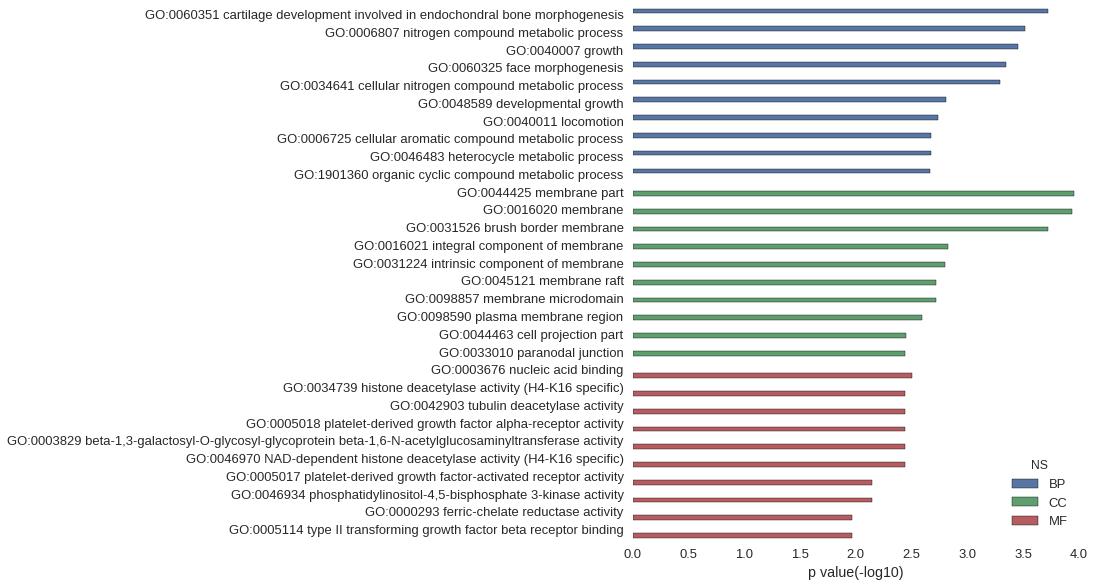
| GO | P value |
|---|---|
| GO:0060351 | 1.91e-04 |
| GO:0006807 | 3.08e-04 |
| GO:0040007 | 3.54e-04 |
| GO:0060325 | 4.56e-04 |
| GO:0034641 | 5.12e-04 |
| GO:0048589 | 1.55e-03 |
| GO:0040011 | 1.83e-03 |
| GO:0006725 | 2.11e-03 |
| GO:0046483 | 2.12e-03 |
| GO:1901360 | 2.18e-03 |
| GO:0044425 | 1.10e-04 |
| GO:0016020 | 1.16e-04 |
| GO:0031526 | 1.91e-04 |
| GO:0016021 | 1.50e-03 |
| GO:0031224 | 1.58e-03 |
| GO:0045121 | 1.90e-03 |
| GO:0098857 | 1.90e-03 |
| GO:0098590 | 2.54e-03 |
| GO:0044463 | 3.59e-03 |
| GO:0033010 | 3.62e-03 |
| GO:0003676 | 3.13e-03 |
| GO:0034739 | 3.62e-03 |
| GO:0042903 | 3.62e-03 |
| GO:0005018 | 3.62e-03 |
| GO:0003829 | 3.62e-03 |
| GO:0046970 | 3.62e-03 |
| GO:0005017 | 7.24e-03 |
| GO:0046934 | 7.24e-03 |
| GO:0000293 | 1.08e-02 |
| GO:0005114 | 1.08e-02 |
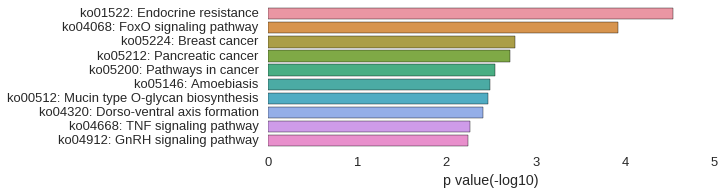
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06644
AGAGTTCTCGTATTCAAAATAAGCCACCGCACACCAAAGCCAGGCAAAACAGCGTGGCCATGAAGGAACG
TTTACCTTGCACAGTCGGTGCCTTTATCAGTATTACAACCTTATTATTAGTACCGTTTTCGCTCCTTTGA
TATCTGAAACTGCTTTGATCTGAAGTGTGTTACTTATTTGTTCTTTTTAGGCACCTTACTTTTTTGTGCA
GTTCACAAATGTGAGTGGAGAACGTCGTTTTTAACCATCCAGTGTTTCTCTGAGCTCTTCAGAAAAAGCA
CACACACAAAAG
AGAGTTCTCGTATTCAAAATAAGCCACCGCACACCAAAGCCAGGCAAAACAGCGTGGCCATGAAGGAACG
TTTACCTTGCACAGTCGGTGCCTTTATCAGTATTACAACCTTATTATTAGTACCGTTTTCGCTCCTTTGA
TATCTGAAACTGCTTTGATCTGAAGTGTGTTACTTATTTGTTCTTTTTAGGCACCTTACTTTTTTGTGCA
GTTCACAAATGTGAGTGGAGAACGTCGTTTTTAACCATCCAGTGTTTCTCTGAGCTCTTCAGAAAAAGCA
CACACACAAAAG