ZFLNCG06647
Basic Information
Chromesome: chr11
Start: 11460182
End: 11460476
Transcript: ZFLNCT10191
Known as: ENSDARG00000088963
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800037 | egg | normal | 6290.81 |
| SRR800043 | sphere stage | normal | 3328.10 |
| SRR800046 | sphere stage | 5azaCyD treatment | 2682.72 |
| SRR800049 | sphere stage | control treatment | 2126.01 |
| SRR800045 | muscle | normal | 110.89 |
| SRR748488 | sperm | normal | 16.43 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 16.16 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 11.93 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 8.48 |
| SRR658539 | bud | Gata5/6 morphant | 5.97 |
Express in tissues
Correlated coding gene
Download
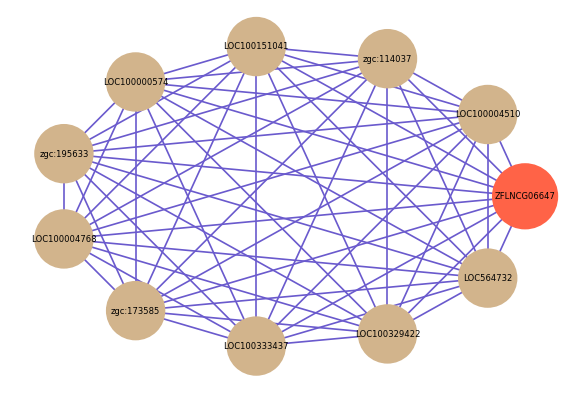
| gene | correlation coefficent |
|---|---|
| LOC100004510 | 0.61 |
| zgc:114037 | 0.60 |
| LOC100151041 | 0.60 |
| LOC100000574 | 0.59 |
| zgc:195633 | 0.59 |
| LOC100004768 | 0.59 |
| zgc:173585 | 0.59 |
| LOC100333437 | 0.58 |
| LOC100329422 | 0.58 |
| LOC564732 | 0.58 |
Gene Ontology
Download
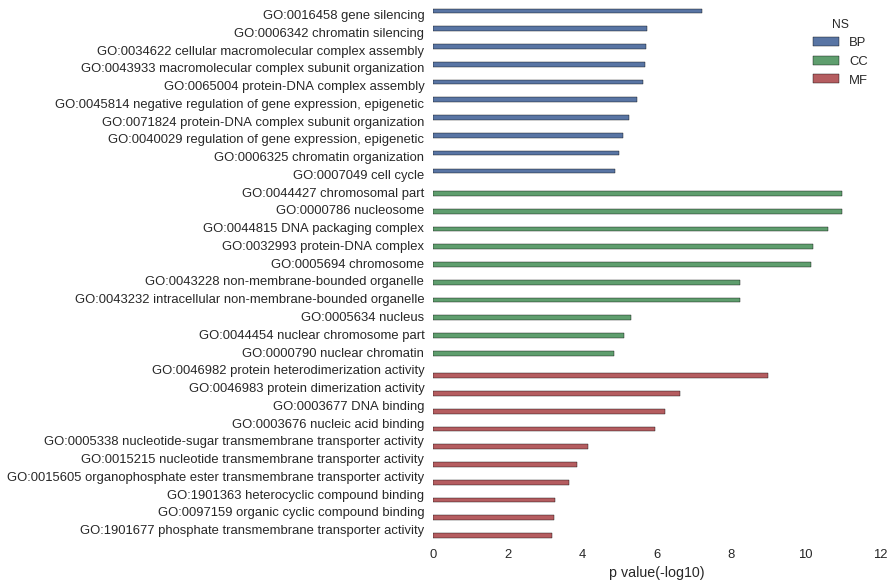
| GO | P value |
|---|---|
| GO:0016458 | 6.21e-08 |
| GO:0006342 | 1.83e-06 |
| GO:0034622 | 1.95e-06 |
| GO:0043933 | 2.05e-06 |
| GO:0065004 | 2.39e-06 |
| GO:0045814 | 3.42e-06 |
| GO:0071824 | 5.44e-06 |
| GO:0040029 | 8.05e-06 |
| GO:0006325 | 1.00e-05 |
| GO:0007049 | 1.32e-05 |
| GO:0044427 | 1.03e-11 |
| GO:0000786 | 1.06e-11 |
| GO:0044815 | 2.47e-11 |
| GO:0032993 | 6.30e-11 |
| GO:0005694 | 7.01e-11 |
| GO:0043228 | 5.58e-09 |
| GO:0043232 | 5.58e-09 |
| GO:0005634 | 5.00e-06 |
| GO:0044454 | 7.55e-06 |
| GO:0000790 | 1.42e-05 |
| GO:0046982 | 1.02e-09 |
| GO:0046983 | 2.33e-07 |
| GO:0003677 | 5.83e-07 |
| GO:0003676 | 1.12e-06 |
| GO:0005338 | 6.88e-05 |
| GO:0015215 | 1.37e-04 |
| GO:0015605 | 2.28e-04 |
| GO:1901363 | 5.26e-04 |
| GO:0097159 | 5.80e-04 |
| GO:1901677 | 6.32e-04 |
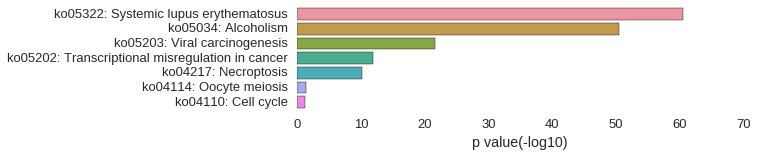
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06647
GTCTGCGTCTCTCTGTGTTGCTCAGGCTGAACTGCAGTGTCTATTCACAGGCGCGATCCCACTACTGATC
AGCACGGGGGCTTTGACCTGCTCCGTCTCCGGCCTGGGACGGTTCACCCCTCCTTAGACGACCTGGTGGT
CCCGAGCTCACCCAGGAGCACCATATCGATACCGAACTTAGTGCGGACACCCGATCGACATCGTCCACTG
CAGCCCAGAAGCCCTGAGCTCAAGCGATCCGCAGACTCAGCCTCCTAGCAGCTTGGATTACAGGCGCGCG
CCACTGAACCCGGC
GTCTGCGTCTCTCTGTGTTGCTCAGGCTGAACTGCAGTGTCTATTCACAGGCGCGATCCCACTACTGATC
AGCACGGGGGCTTTGACCTGCTCCGTCTCCGGCCTGGGACGGTTCACCCCTCCTTAGACGACCTGGTGGT
CCCGAGCTCACCCAGGAGCACCATATCGATACCGAACTTAGTGCGGACACCCGATCGACATCGTCCACTG
CAGCCCAGAAGCCCTGAGCTCAAGCGATCCGCAGACTCAGCCTCCTAGCAGCTTGGATTACAGGCGCGCG
CCACTGAACCCGGC