ZFLNCG06663
Basic Information
Chromesome: chr11
Start: 11548256
End: 11548550
Transcript: ZFLNCT10207
Known as: ENSDARG00000090123
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800043 | sphere stage | normal | 3769.45 |
| SRR800046 | sphere stage | 5azaCyD treatment | 3253.46 |
| SRR800037 | egg | normal | 2621.34 |
| SRR800049 | sphere stage | control treatment | 1831.38 |
| SRR800045 | muscle | normal | 52.81 |
| SRR748488 | sperm | normal | 33.71 |
| SRR801555 | embryo | RPS19 morpholino | 7.76 |
| SRR658539 | bud | Gata5/6 morphant | 7.37 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 6.23 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 4.74 |
Express in tissues
Correlated coding gene
Download
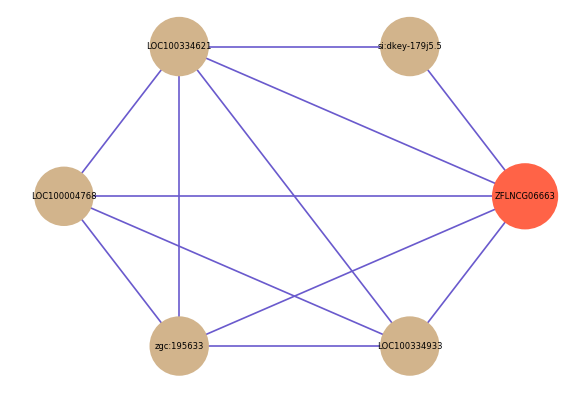
| gene | correlation coefficent |
|---|---|
| si:dkey-179j5.5 | 0.53 |
| LOC100334621 | 0.52 |
| LOC100004768 | 0.52 |
| zgc:195633 | 0.50 |
| LOC100334933 | 0.50 |
Gene Ontology
Download
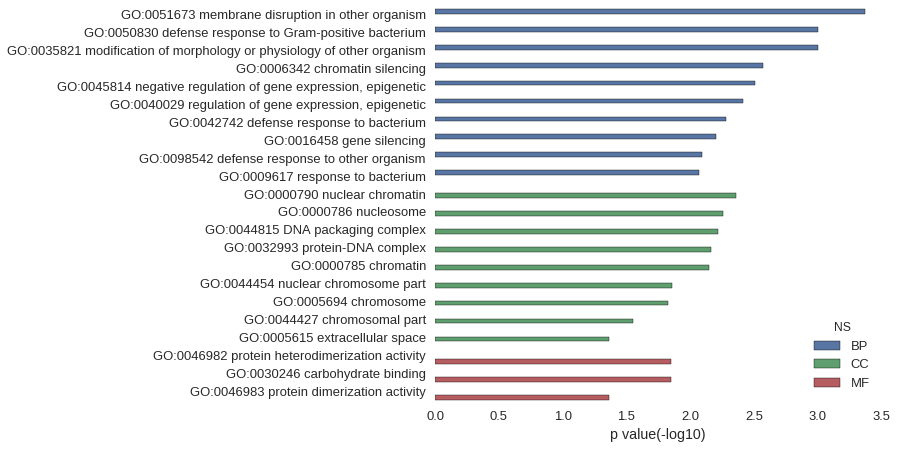
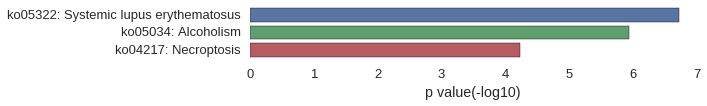
| GO | P value |
|---|---|
| GO:0051673 | 4.26e-04 |
| GO:0050830 | 9.95e-04 |
| GO:0035821 | 9.95e-04 |
| GO:0006342 | 2.70e-03 |
| GO:0045814 | 3.12e-03 |
| GO:0040029 | 3.83e-03 |
| GO:0042742 | 5.25e-03 |
| GO:0016458 | 6.24e-03 |
| GO:0098542 | 8.09e-03 |
| GO:0009617 | 8.51e-03 |
| GO:0000790 | 4.40e-03 |
| GO:0000786 | 5.54e-03 |
| GO:0044815 | 6.10e-03 |
| GO:0032993 | 6.81e-03 |
| GO:0000785 | 7.09e-03 |
| GO:0044454 | 1.39e-02 |
| GO:0005694 | 1.49e-02 |
| GO:0044427 | 2.82e-02 |
| GO:0005615 | 4.30e-02 |
| GO:0046982 | 1.40e-02 |
| GO:0030246 | 1.42e-02 |
| GO:0046983 | 4.33e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06663
GTCTGCGTCTCTCTGTGTCGCTCAGGCTGCACTGCAGTGTCTATTCACAGGCGCGATCCCACTACTGATC
AGCACGGGGGCTTTGACCTGCTCCGTGTCCGGCCTGGGTCGGTTCACCCCTTCTTAAACGACCTGGTGGT
CCCAAGCTCCCCCAGCAGCACCATATCGATACCGAACTTAACGTGGACACCCGATCAACATAGTCCACTG
CAGTCCAGAAGCCCTGAACTCAAGCGATCCGCAGCCTCAGCCTCCCAGTAACTTGGATTACAGGCGCGCG
CCACTGAACCCGGC
GTCTGCGTCTCTCTGTGTCGCTCAGGCTGCACTGCAGTGTCTATTCACAGGCGCGATCCCACTACTGATC
AGCACGGGGGCTTTGACCTGCTCCGTGTCCGGCCTGGGTCGGTTCACCCCTTCTTAAACGACCTGGTGGT
CCCAAGCTCCCCCAGCAGCACCATATCGATACCGAACTTAACGTGGACACCCGATCAACATAGTCCACTG
CAGTCCAGAAGCCCTGAACTCAAGCGATCCGCAGCCTCAGCCTCCCAGTAACTTGGATTACAGGCGCGCG
CCACTGAACCCGGC