ZFLNCG06672
Basic Information
Chromesome: chr11
Start: 11582182
End: 11582476
Transcript: ZFLNCT10216
Known as: ENSDARG00000086852
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800046 | sphere stage | 5azaCyD treatment | 1705.13 |
| SRR800043 | sphere stage | normal | 1401.43 |
| SRR800049 | sphere stage | control treatment | 1248.82 |
| SRR800037 | egg | normal | 987.17 |
| SRR800045 | muscle | normal | 110.89 |
| SRR535986 | larvae | nhsl1b mut | 8.36 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 7.02 |
| SRR527844 | 24hpf | normal | 4.18 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 2.81 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 2.80 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| LOC565028 | 0.58 |
| LOC100329422 | 0.56 |
| LOC565202 | 0.56 |
| LOC565341 | 0.56 |
| LOC100000930 | 0.56 |
| si:ch211-113a14.16 | 0.55 |
| LOC563490 | 0.55 |
| LOC100333437 | 0.54 |
| LOC100002181 | 0.54 |
| si:ch211-113a14.28 | 0.54 |
Gene Ontology
Download
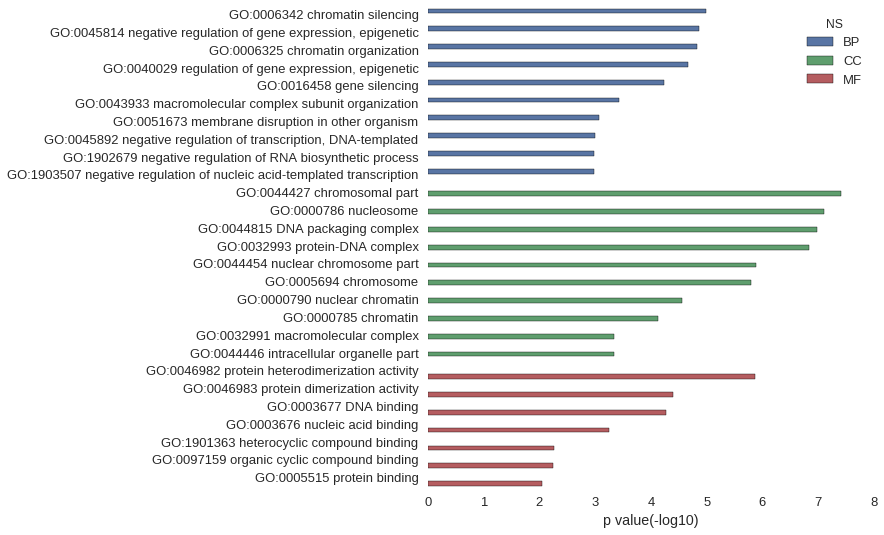
| GO | P value |
|---|---|
| GO:0006342 | 1.03e-05 |
| GO:0045814 | 1.40e-05 |
| GO:0006325 | 1.47e-05 |
| GO:0040029 | 2.12e-05 |
| GO:0016458 | 5.71e-05 |
| GO:0043933 | 3.75e-04 |
| GO:0051673 | 8.53e-04 |
| GO:0045892 | 1.02e-03 |
| GO:1902679 | 1.04e-03 |
| GO:1903507 | 1.04e-03 |
| GO:0044427 | 3.96e-08 |
| GO:0000786 | 7.86e-08 |
| GO:0044815 | 1.06e-07 |
| GO:0032993 | 1.49e-07 |
| GO:0044454 | 1.30e-06 |
| GO:0005694 | 1.61e-06 |
| GO:0000790 | 2.81e-05 |
| GO:0000785 | 7.39e-05 |
| GO:0032991 | 4.58e-04 |
| GO:0044446 | 4.61e-04 |
| GO:0046982 | 1.34e-06 |
| GO:0046983 | 4.09e-05 |
| GO:0003677 | 5.35e-05 |
| GO:0003676 | 5.69e-04 |
| GO:1901363 | 5.52e-03 |
| GO:0097159 | 5.74e-03 |
| GO:0005515 | 9.04e-03 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06672
CCGGGTTCAGTGGCGCGCGCCTGTAATCCAAGTTACTAGGAGGCTGAGTCTGCGGATCGCTTGAGCTCAG
GGCTTCTGGACTGCAGTGTATTATGTCGATCGGGAGTCCGCACTAAGTTCAGTGTCGATATGGTGCTCCT
GAGGGAGCTCAGGATCACCAGGTCGTCTAAAGAGGGGTGAACCGGCCCAGGTCGGAGACGGAGCAGGTCA
AAGCCCCCGTGCTGATCAGTAGTGGGTTCGCACCTGTGAATAGACACTGCAGTGCAGCCTGAGCGACACA
GAGAGACGCAGACT
CCGGGTTCAGTGGCGCGCGCCTGTAATCCAAGTTACTAGGAGGCTGAGTCTGCGGATCGCTTGAGCTCAG
GGCTTCTGGACTGCAGTGTATTATGTCGATCGGGAGTCCGCACTAAGTTCAGTGTCGATATGGTGCTCCT
GAGGGAGCTCAGGATCACCAGGTCGTCTAAAGAGGGGTGAACCGGCCCAGGTCGGAGACGGAGCAGGTCA
AAGCCCCCGTGCTGATCAGTAGTGGGTTCGCACCTGTGAATAGACACTGCAGTGCAGCCTGAGCGACACA
GAGAGACGCAGACT

