ZFLNCG06691
Basic Information
Chromesome: chr11
Start: 12285809
End: 12286103
Transcript: ZFLNCT10253
Known as: ENSDARG00000090964
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800046 | sphere stage | 5azaCyD treatment | 1787.07 |
| SRR800043 | sphere stage | normal | 1420.99 |
| SRR800049 | sphere stage | control treatment | 1381.63 |
| SRR800037 | egg | normal | 524.71 |
| SRR800045 | muscle | normal | 351.16 |
| SRR801556 | embryo | RPS19 and p53 morpholino | 71.16 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 35.62 |
| SRR748488 | sperm | normal | 22.33 |
| SRR527844 | 24hpf | normal | 21.20 |
| SRR801555 | embryo | RPS19 morpholino | 21.05 |
Express in tissues
Correlated coding gene
Download
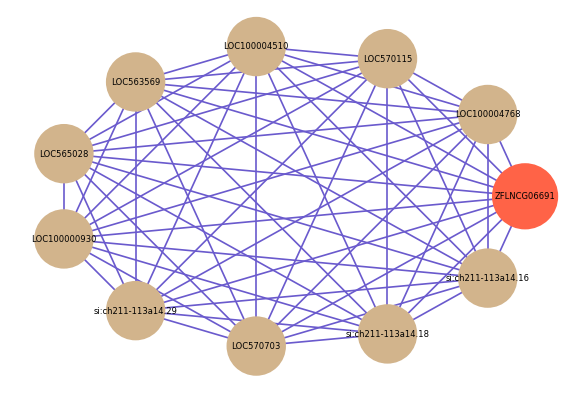
| gene | correlation coefficent |
|---|---|
| LOC100004768 | 0.65 |
| LOC570115 | 0.62 |
| LOC100004510 | 0.61 |
| LOC563569 | 0.60 |
| LOC565028 | 0.60 |
| LOC100000930 | 0.60 |
| si:ch211-113a14.29 | 0.60 |
| LOC570703 | 0.59 |
| si:ch211-113a14.18 | 0.59 |
| si:ch211-113a14.16 | 0.59 |
Gene Ontology
Download
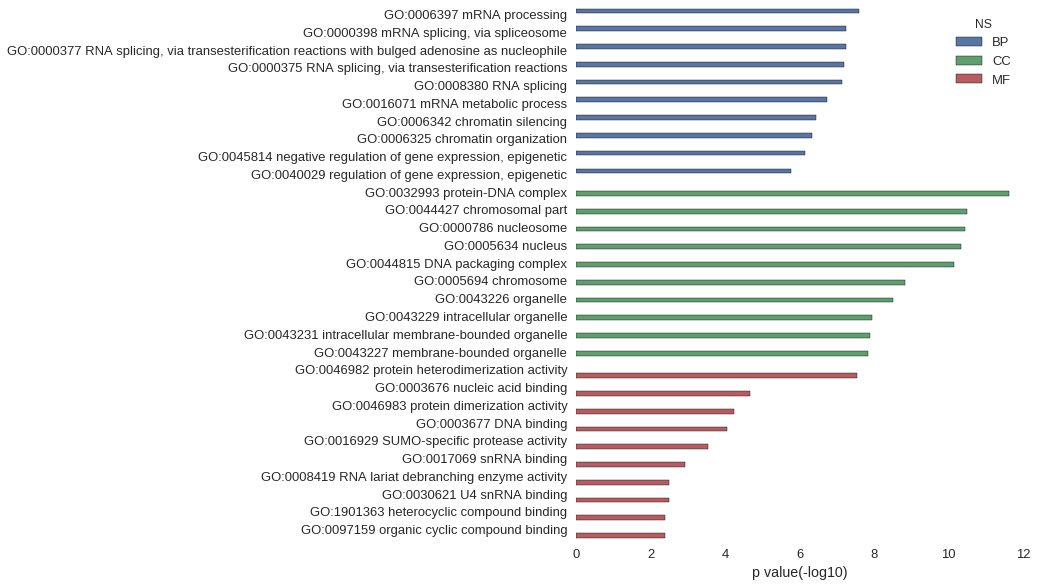
| GO | P value |
|---|---|
| GO:0006397 | 2.56e-08 |
| GO:0000398 | 5.66e-08 |
| GO:0000377 | 5.66e-08 |
| GO:0000375 | 6.22e-08 |
| GO:0008380 | 7.38e-08 |
| GO:0016071 | 1.81e-07 |
| GO:0006342 | 3.74e-07 |
| GO:0006325 | 4.75e-07 |
| GO:0045814 | 7.00e-07 |
| GO:0040029 | 1.66e-06 |
| GO:0032993 | 2.35e-12 |
| GO:0044427 | 3.14e-11 |
| GO:0000786 | 3.52e-11 |
| GO:0005634 | 4.65e-11 |
| GO:0044815 | 7.30e-11 |
| GO:0005694 | 1.51e-09 |
| GO:0043226 | 3.02e-09 |
| GO:0043229 | 1.14e-08 |
| GO:0043231 | 1.26e-08 |
| GO:0043227 | 1.48e-08 |
| GO:0046982 | 2.94e-08 |
| GO:0003676 | 2.22e-05 |
| GO:0046983 | 5.78e-05 |
| GO:0003677 | 8.69e-05 |
| GO:0016929 | 2.89e-04 |
| GO:0017069 | 1.22e-03 |
| GO:0008419 | 3.27e-03 |
| GO:0030621 | 3.27e-03 |
| GO:1901363 | 4.09e-03 |
| GO:0097159 | 4.22e-03 |
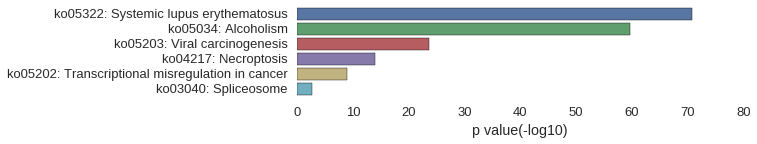
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06691
GTCTGCATCTCTCTGTGTAGCTCAGGCTGCACTGCAGTGTCTATTCACAGGCGCGATCCCACTACTGATC
GGCACGGGGGCTTTGACCTGCTTCGTCTCCGACCTGGGCCGGTTCACCCCTCTTTACACGACCTGGTGGT
CCCGAGCTCCCCAAGGAGCACCATATCGATACCGAACTTAGTGCGGACACCCGATCGACATAGTCCACTG
CAGCCCAGAAGCCCTGAGCTCAAGCGATCCGCAGCCTCAGCCTCCCAGTAGCTTGGATTACAGGCGCGCG
CCACTGAACCCGGC
GTCTGCATCTCTCTGTGTAGCTCAGGCTGCACTGCAGTGTCTATTCACAGGCGCGATCCCACTACTGATC
GGCACGGGGGCTTTGACCTGCTTCGTCTCCGACCTGGGCCGGTTCACCCCTCTTTACACGACCTGGTGGT
CCCGAGCTCCCCAAGGAGCACCATATCGATACCGAACTTAGTGCGGACACCCGATCGACATAGTCCACTG
CAGCCCAGAAGCCCTGAGCTCAAGCGATCCGCAGCCTCAGCCTCCCAGTAGCTTGGATTACAGGCGCGCG
CCACTGAACCCGGC